与 Cas9 核酸酶相比,碱基编辑器和质点编辑器在 DNA 修复过程中发生 DNA 大缺失的频率低 20 倍
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
用于编辑基因组时,Cas9 核酸酶会在 DNA 上产生定向双链断裂。随后的DNA修复途径会诱发大的基因组缺失(大于100 bp),这限制了基因组编辑的适用性。我们在这里发现,Cas9 介导的双链断裂会在癌细胞系、人类胚胎干细胞和人类原代 T 细胞中以不同的频率诱导大的缺失,而且大多数缺失是通过两种修复途径产生的:末端切除和 DNA 聚合酶 theta 介导的末端连接。这些发现要求优化长程扩增片段测序、开发用于同时分析大DNA缺失和小DNA改变的k-mer比对算法,以及使用CRISPR干扰筛选。尽管利用突变的Cas9缺口酶会产生单链断裂,碱基编辑器和质粒编辑器也会产生大的缺失,但频率比Cas9低约20倍。我们提供了减少这种缺失的策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。
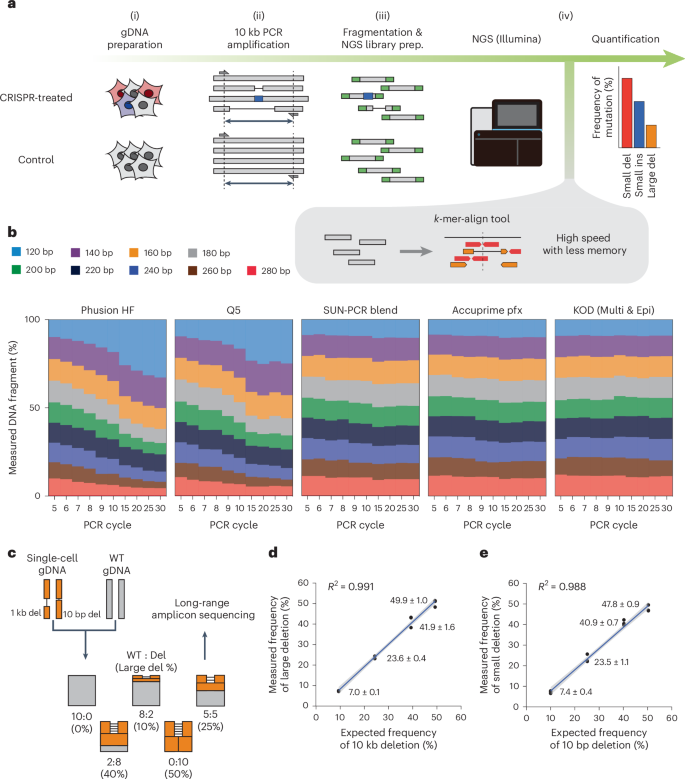

Large DNA deletions occur during DNA repair at 20-fold lower frequency for base editors and prime editors than for Cas9 nucleases
When used to edit genomes, Cas9 nucleases produce targeted double-strand breaks in DNA. Subsequent DNA-repair pathways can induce large genomic deletions (larger than 100 bp), which constrains the applicability of genome editing. Here we show that Cas9-mediated double-strand breaks induce large deletions at varying frequencies in cancer cell lines, human embryonic stem cells and human primary T cells, and that most deletions are produced by two repair pathways: end resection and DNA-polymerase theta-mediated end joining. These findings required the optimization of long-range amplicon sequencing, the development of a k-mer alignment algorithm for the simultaneous analysis of large DNA deletions and small DNA alterations, and the use of CRISPR-interference screening. Despite leveraging mutated Cas9 nickases that produce single-strand breaks, base editors and prime editors also generated large deletions, yet at approximately 20-fold lower frequency than Cas9. We provide strategies for the mitigation of such deletions. DNA repair after Cas9-mediated double-strand breaks induces large DNA deletions at frequencies 20-fold higher than elicited by base editors and prime editors leveraging Cas9 nickases producing single-strand breaks.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


