针对 SARS CoV-2 RNA 依赖性 RNA 聚合酶的 AYUSH-64 药用植物植物化合物的硅学探索。
IF 1.9
Q3 INTEGRATIVE & COMPLEMENTARY MEDICINE
引用次数: 0
摘要
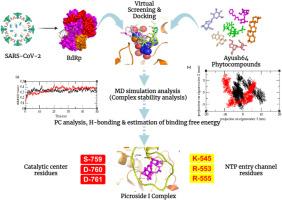
背景:AYUSH 64 配方有助于治疗轻度至中度的 COVID-19 病例。虽然已经提出了几种抗 COVID-19 的药物,但还没有治疗 SARS-CoV-2 感染的药物。RNA 依赖性 RNA 聚合酶(RdRp)是 SARS-CoV-2 复制的关键酶,因此可将其视为实验研究中更好的药物靶点:AYUSH-64 配方植物具有多种治疗特性;因此,本研究旨在筛选这些植物中针对 SARS CoV2 RdRp 的植物化合物,以确定可能影响 COVID-19 感染的特定化合物:使用 PatchDock 和 AutoDock 工具进行对接实验。分别在 GROMACS v2019.4 和 Gaussian 09 软件中对蛋白质配体 Picroside-I 和 Remdesivir 复合物进行了 MD 模拟和密度泛函理论(DFT)计算:在测试的植物化合物中,来自AYUSH-64药用植物的五种植物化合物(Picroside I、齐墩果酸、Arvenin I、II和III)显示可能与RdRp催化残基(Ser759、Asp760和Asp761)结合。其中,Picroside I 与 NTP 进入通道残基(Arg553 和 Arg555)有氢键相互作用。MM-PBSA 自由能、RMSD、Rg、PCA 和 RMSF 分析表明,在 50 ns 模拟中,Picroside I 复合物与 RdRp 发生了稳定的结合相互作用。此外,经 DFT 证实,Picroside I 对目标蛋白质具有稳健的吸引力:本研究结果表明,AYUSH 64 种药用植物化合物中的 Picroside I 能选择性地结合 SARS-CoV2 RdRp 的催化和 NTP 进入通道残基,因此可被视为 SARS-CoV2 RdRp 的潜在抑制剂。本文章由计算机程序翻译,如有差异,请以英文原文为准。
In silico exploration of phytocompounds from AYUSH-64 medicinal plants against SARS CoV-2 RNA-dependent RNA polymerase
Background
The AYUSH 64 formulation helps to treat mild to moderate cases of COVID-19. Although several drugs have been proposed to combat COVID-19, no medication is available for SARS-CoV-2 infection. The RNA-dependent RNA polymerase (RdRp) is the pivotal enzyme of SARS-CoV-2 replication, so it could be considered a better drug target for experimental studies.
Objective
The AYUSH-64 formulation plants exhibited multiple therapeutic properties; thus, the present study aims to screen the phytocompounds of these plants against SARS CoV2 RdRp to identify specific compounds that could potentially affect COVID-19 infection.
Materials and methods
PatchDock and AutoDock tools were used for docking experiments. MD simulations and Density Functional Theory (DFT) calculations of protein-ligand Picroside-I and Remdesivir complexes were carried out in GROMACS v2019.4 and Gaussian 09 software, respectively.
Results
Among the tested, five phytocompounds (Picroside I, Oleanolic acid, Arvenin I, II, and III) from AYUSH-64 medicinal plants showed possible binding with RdRp catalytic residues (Ser759, Asp760, and Asp761). Of these, Picroside I exhibited hydrogen bond interactions with NTP entry channel residues (Arg553 and Arg555). The MM-PBSA free energy, RMSD, Rg, PCA, and RMSF analysis suggested that the Picroside I complex showed stable binding interactions with RdRp in the 50 ns simulation. In addition to this, Picroside I revealed its robust and attractive nature toward the target protein, as confirmed by DFT.
Conclusion
The results of this study have proposed that Picroside I from AYUSH 64 medicinal plant compounds was the selective binder of catalytic and NTP entry channel residues of SARS-CoV2 RdRp thereby; it may considered as a potential inhibitor of SARS-CoV2 RdRp.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Ayurveda and Integrative Medicine
INTEGRATIVE & COMPLEMENTARY MEDICINE-
CiteScore
4.70
自引率
12.50%
发文量
136
审稿时长
30 weeks
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


