拓扑 DNA 混合物表现出共振形变场和应变传播动力学,并通过立体约束进行调整。
IF 9.4
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
了解聚合物如何因局部应力和应变而变形,以及应变如何从局部扰动中传播,是材料制造到细胞力学等广泛领域面临的巨大挑战。对于具有不同拓扑结构的聚合物混合物而言,这些动态变化尤为复杂,其中可能存在多种不同的物种依赖机制。在这里,我们使用 OpTiDDM(光学镊子集成差分动态显微镜)来阐明不同大小的线性、环状和超卷曲 DNA 二元共混物的变形场和传播动力学。我们揭示了应变排列和超扩散传输与应变速率的非单调依赖性。然而,峰值排列和超扩散性却出人意料地解耦了,它们出现在不同的应变速率下,与不同拓扑结构的不同弛豫速率产生共振。尽管存在这种普遍共振,但我们发现环状线性混合物的应变传播是由缠结决定的,而超螺旋环状混合物则是由劳斯动力学决定的。我们的研究结果捕捉到了拓扑混合物在传播和变形动力学方面的关键微妙之处,揭示了新的物理规律,并为实现响应特征的解耦调整提供了一条途径。我们预计,我们的方法可广泛用于绘制聚合物共混物的变形动力学图,从而实现自下而上的定制材料设计。意义声明:在生物学和制造业中,生物材料经常受到局部和空间不均匀应变和应力的影响。然而,了解应变在不同时空尺度上的吸收、分布或传播程度仍然是一个巨大的挑战。在这里,我们将光学镊子与差分动态显微镜相结合,阐明了线性、环状和超螺旋 DNA 混合体的形变场和传播动力学,揭示了应变排列和超扩散性的稳健非单调趋势和解耦,并捕捉到了传播和形变动力学中的关键微妙之处。我们的研究结果为指导响应特征的解耦调整提供了新的重要物理视角,可用于绘制各种生物聚合物和其他大分子系统的变形动力学图,从而实现自下而上的定制生物材料设计。本文章由计算机程序翻译,如有差异,请以英文原文为准。
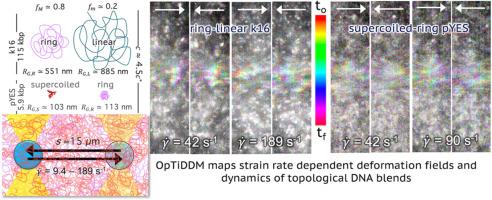
Topological DNA blends exhibit resonant deformation fields and strain propagation dynamics tuned by steric constraints
Understanding how polymers deform in response to local stresses and strains, and how strains propagate from a local disturbance, are grand challenges in wide-ranging fields from materials manufacturing to cell mechanics. These dynamics are particularly complex for blends of polymers of distinct topologies, for which several different species-dependent mechanisms may contribute. Here, we use OpTiDDM (Optical Tweezers integrating Differential Dynamic Microscopy) to elucidate deformation fields and propagation dynamics of binary blends of linear, ring and supercoiled DNA of varying sizes. We reveal robust non-monotonic dependence of strain alignment and superdiffusive transport with strain rate. However, peak alignment and superdiffusivity are surprisingly decoupled, occurring at different strain rates resonant with the distinct relaxation rates of the different topologies. Despite this universal resonance, we find that strain propagation of ring-linear blends is dictated by entanglements while supercoiled-ring blends are governed by Rouse dynamics. Our results capture critical subtleties in propagation and deformation dynamics of topological blends, shedding new light on the governing physics and offering a route towards decoupled tuning of response features. We anticipate our approach to be broadly generalizable to mapping the deformation dynamics of polymer blends, with an eye towards bottom-up bespoke materials design.
Statement of Significance
In biology and in manufacturing, biomaterials are often subject to localized and spatially nonuniform strains and stresses. Yet, understanding the extent to which strains are absorbed, distributed, or propagated across different spatiotemporal scales remains a grand challenge. Here, we combine optical tweezers with differential dynamic microscopy to elucidate deformation fields and propagation dynamics of blends of linear, ring and supercoiled DNA, revealing robust non-monotonic trends and decoupling of strain alignment and superdiffusivity, and capturing critical subtleties in propagation and deformation dynamics. Our results, shedding important new physical insight to guide decoupled tuning of response features, may be leveraged to map the deformation dynamics of wide-ranging systems of biopolymers and other macromolecules, with an eye towards bottom-up bespoke biomaterials design.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Acta Biomaterialia
工程技术-材料科学:生物材料
CiteScore
16.80
自引率
3.10%
发文量
776
审稿时长
30 days
期刊介绍:
Acta Biomaterialia is a monthly peer-reviewed scientific journal published by Elsevier. The journal was established in January 2005. The editor-in-chief is W.R. Wagner (University of Pittsburgh). The journal covers research in biomaterials science, including the interrelationship of biomaterial structure and function from macroscale to nanoscale. Topical coverage includes biomedical and biocompatible materials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


