仅利用基因组信息预测整个埃希氏菌属的菌株级噬菌体-宿主相互作用
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
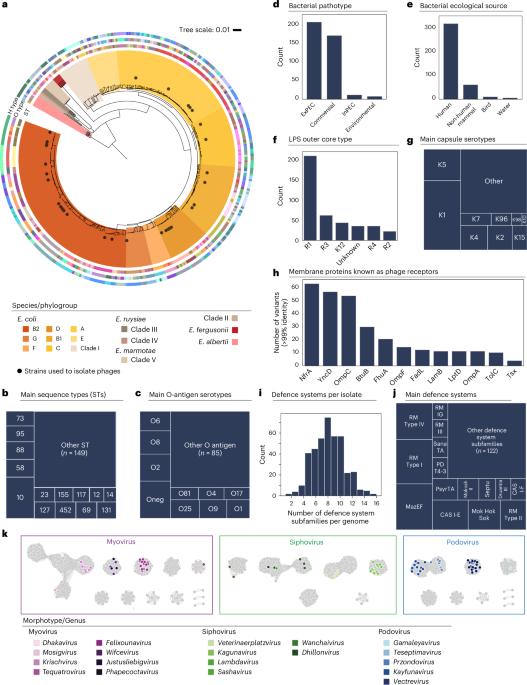
预测噬菌体感染特定细菌菌株有望推动噬菌体疗法和微生物生态学的发展。目前尚不清楚成熟的噬菌体-宿主模型系统的动态是否适用于种类繁多的微生物。在这里,我们展示了仅利用基因组数据就能准确预测埃希氏菌属天然分离株中噬菌体-细菌相互作用的结果(接收者操作特征曲线下面积(AUROC)为 86%)。我们通过实验建立了 403 种不同的埃希氏菌株与 96 种噬菌体之间的相互作用数据集。大多数相互作用都是由吸附因素引起的,而抗噬菌体系统的作用则微乎其微。我们训练了预测算法,并指出了预测不准确的相互作用,以指导未来的研究工作。最后,我们建立了一个管道来推荐量身定制的噬菌体鸡尾酒,并在 100 个致病性大肠杆菌分离物上证明了其效率。这项工作提供了对噬菌体宿主特异性的定量见解,并支持在噬菌体疗法中使用预测算法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Prediction of strain level phage–host interactions across the Escherichia genus using only genomic information
Predicting bacteriophage infection of specific bacterial strains promises advancements in phage therapy and microbial ecology. Whether the dynamics of well-established phage–host model systems generalize to the wide diversity of microbes is currently unknown. Here we show that we could accurately predict the outcomes of phage–bacteria interactions at the strain level in natural isolates from the genus Escherichia using only genomic data (area under the receiver operating characteristic curve (AUROC) of 86%). We experimentally established a dataset of interactions between 403 diverse Escherichia strains and 96 phages. Most interactions are explained by adsorption factors as opposed to antiphage systems which play a marginal role. We trained predictive algorithms and pinpoint poorly predicted interactions to direct future research efforts. Finally, we established a pipeline to recommend tailored phage cocktails, demonstrating efficiency on 100 pathogenic E. coli isolates. This work provides quantitative insights into phage–host specificity and supports the use of predictive algorithms in phage therapy. Phage–host interactions are computationally predicted using only genomic information, highlighting future research directions and enabling generation of custom phage cocktails.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


