利用区域形态学多重图像标记(MILWRM)在空间 omics 数据中进行共识组织域检测。
IF 5.2
1区 生物学
Q1 BIOLOGY
引用次数: 0
摘要
空间分辨分子测定法提供了原位和各种分辨率的高维遗传、转录组、蛋白质组和表观遗传信息。将这些跨模式数据与组织学特征配对,可以在完整的微环境和组织结构背景下对组织病理学进行强有力的研究。分子分析物和样本的维度不断增加,需要新的数据科学方法对空间解析的分子数据进行功能注释。一个具体的挑战是数据驱动的跨样本领域检测,以便在组织图谱绘制工作产生的大量多重数据集上,对共识组织区划内和组织区划间进行分析。在此,我们介绍了 MILWRM(区域形态多重图像标记)--一个 Python 软件包,用于在图像或斑点级别快速进行多尺度组织域检测和注释。我们展示了 MILWRM 在人类结肠息肉、淋巴结、小鼠肾脏和小鼠大脑切片中,通过来自不同平台的两种不同空间数据模式的空间信息聚类,识别组织学上不同分区的实用性。我们利用在人类结肠息肉中检测到的组织域来阐明息肉亚型之间的分子区别,并探索了 MILWRM 识别脑组织解剖区域及其各自不同分子特征的能力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
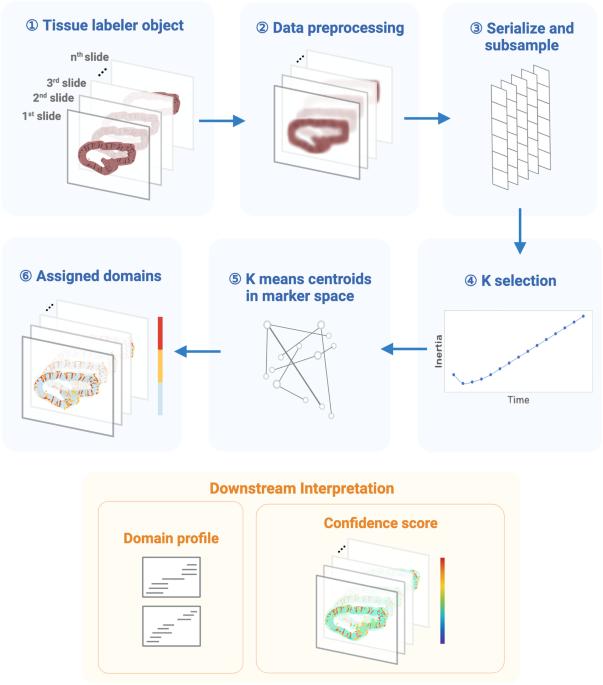
Consensus tissue domain detection in spatial omics data using multiplex image labeling with regional morphology (MILWRM)
Spatially resolved molecular assays provide high dimensional genetic, transcriptomic, proteomic, and epigenetic information in situ and at various resolutions. Pairing these data across modalities with histological features enables powerful studies of tissue pathology in the context of an intact microenvironment and tissue structure. Increasing dimensions across molecular analytes and samples require new data science approaches to functionally annotate spatially resolved molecular data. A specific challenge is data-driven cross-sample domain detection that allows for analysis within and between consensus tissue compartments across high volumes of multiplex datasets stemming from tissue atlasing efforts. Here, we present MILWRM (multiplex image labeling with regional morphology)—a Python package for rapid, multi-scale tissue domain detection and annotation at the image- or spot-level. We demonstrate MILWRM’s utility in identifying histologically distinct compartments in human colonic polyps, lymph nodes, mouse kidney, and mouse brain slices through spatially-informed clustering in two different spatial data modalities from different platforms. We used tissue domains detected in human colonic polyps to elucidate the molecular distinction between polyp subtypes, and explored the ability of MILWRM to identify anatomical regions of the brain tissue and their respective distinct molecular profiles. MILWRM is a Python package that can perform tissue domain detection on spatial transcriptomics (ST) and multiplex immunofluorescence (mIF) data across multiple specimens.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


