组合转录因子结合编码小鼠前脑GABA能神经发生的顺式调控线路
IF 10.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
转录因子(TF)与顺式调控元件组合结合,协调转录程序。尽管对染色质状态和染色体相互作用的研究已经展示了动态的神经发育顺式调控景观,但对TF相互作用的平行理解却相对滞后。为了阐明驱动小鼠基底节发育的TF组合结合,我们整合了12种TF的染色质免疫沉淀测序(ChIP-seq)、H3K4me3相关的增强子-启动子相互作用、染色质和基因表达数据以及功能增强子测定。我们发现了几组具有共享TF结合的推定调控元件(TF-pRE模块),它们协调GABA能神经发生的不同过程并抑制其他细胞命运。大多数 pRE 被一个或两个 TF 结合;但也有一小部分被广泛结合。这些序列具有特殊的进化保守性和主题密度、复杂的染色体相互作用以及作为体内增强子的活性。我们的研究结果提供了对在端脑神经发生过程中激活和抑制表达程序的组合 TF-pRE 相互作用的见解,并证明了 TF 结合对发育转录线路建模的价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
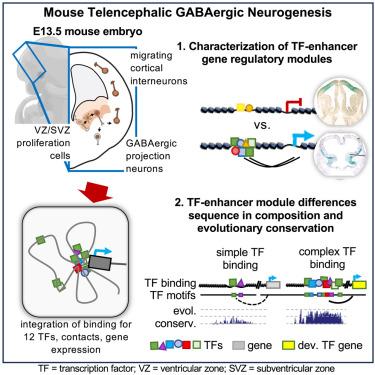
Combinatorial transcription factor binding encodes cis-regulatory wiring of mouse forebrain GABAergic neurogenesis
Transcription factors (TFs) bind combinatorially to cis-regulatory elements, orchestrating transcriptional programs. Although studies of chromatin state and chromosomal interactions have demonstrated dynamic neurodevelopmental cis-regulatory landscapes, parallel understanding of TF interactions lags. To elucidate combinatorial TF binding driving mouse basal ganglia development, we integrated chromatin immunoprecipitation sequencing (ChIP-seq) for twelve TFs, H3K4me3-associated enhancer-promoter interactions, chromatin and gene expression data, and functional enhancer assays. We identified sets of putative regulatory elements with shared TF binding (TF-pRE modules) that orchestrate distinct processes of GABAergic neurogenesis and suppress other cell fates. The majority of pREs were bound by one or two TFs; however, a small proportion were extensively bound. These sequences had exceptional evolutionary conservation and motif density, complex chromosomal interactions, and activity as in vivo enhancers. Our results provide insights into the combinatorial TF-pRE interactions that activate and repress expression programs during telencephalon neurogenesis and demonstrate the value of TF binding toward modeling developmental transcriptional wiring.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


