使用两种生物刺激剂对石油烃污染土壤进行生物修复的实地效果评估
IF 4.1
2区 环境科学与生态学
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
International Biodeterioration & Biodegradation
Pub Date : 2024-10-20
DOI:10.1016/j.ibiod.2024.105942
引用次数: 0
摘要
众所周知,养分是受石油碳氢化合物(PHC)污染的土壤成功进行生物修复的限制因素。因此,我们测试了两种生物刺激剂--土壤生物修复营养素(SBNs)和营养液(NS),以评估它们对石油烃降解的影响,以及在田间使用第三代测序、Tax4Fun2 和 qPCR 对细菌群落组成、功能和功能基因变化的影响。添加 SBN 或 NS 能显著增加并加速总石油烃(TPH)的降解。添加了 SBNs 的生物堆(尤其是 0.5% 的 SBNs)的 TPH 降解率(96%)和 K 值(0.134/天)最高,表明 SBNs 是一种良好的生物刺激剂,可用于对受 PHC 污染的土壤进行生物修复。经过不同的操作后,5 个生物堆的细菌群落和功能迅速重建,代谢功能相对较高,主要的 PHC 降解菌从 Rugosibacter aromaticivorans 和 Novosphingobium aromaticivorans 变为 Immundisolibacter cernigliae 和 Dyella ginsengisoli。Tax4Fun2 预测结果表明,在土壤翻动过程中添加 SBNs 和水可增强和加速细菌的代谢功能以及降解芳香化合物的相关功能基因。qPCR 结果表明,大多数样品中含有较高丰度的 alkB、alkB1、XylE 和 PHE,且 XylE 和 PHE 的表达量较高,而 SBNs 和 NS 的添加增强了 XylE 和 PHE 降解儿茶酚和苯酚的表达量。TPH 和营养物质的浓度是影响 TPH、烷烃和芳香族化合物降解以及细菌群落、功能和功能基因(如 alkB、alkB1、XylE、NAH、PHE 和 BPH4)变化的主要环境因素。事实证明,通过添加 SBNs 进行生物刺激是增加 PHC 降解、促进 PHC 降解菌生长和新陈代谢活动的有效策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。
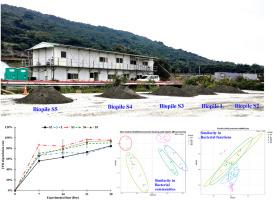
A field-scale assessment of the efficacy of bioremediation of petroleum hydrocarbon-contaminated soils using two biostimulants
Nutrients are known limiting factors for the successful bioremediation of soil contaminated with petroleum hydrocarbons (PHCs). Thus, two biostimulants, soil bioremediation nutrients (SBNs) and nutrient solution (NS), were tested to assess their effects on the degradation of PHCs and changes in bacterial community compositions, functions and functional genes using third generation sequencing, Tax4Fun2 and qPCR at the field scale. The addition of SBNs or NS significantly increased and accelerated the degradation of total petroleum hydrocarbons (TPH). The highest TPH degradation rate (96%) and k (0.134/day) were shown for biopiles with SBNs addition (particularly for 0.5% SBNs), indicating that SBNs were a good biostimulant for bioremediation of PHC-contaminated soil in the field. After different operations, rapid reconstruction of bacterial communities and functions for 5 biopiles was found with relatively high metabolic function and a change in dominant PHC degraders from Rugosibacter aromaticivorans and Novosphingobium aromaticivorans to Immundisolibacter cernigliae and Dyella ginsengisoli. The results of Tax4Fun2 prediction revealed that the addition of SBNs and water with soil turning enhanced and accelerated bacterial metabolic functions and related functional genes for degrading aromatic compounds. The qPCR results indicated that most samples contained a relatively high abundance of alkB, alkB1, XylE and PHE, and high expression of XylE and PHE, while the addition of SBNs and NS enhanced the expression of XylE and PHE for degrading catechol and phenol. Concentrations of TPH and nutrients were major environmental factors influencing the degradation of TPH, alkane and aromatic compounds and changes in bacterial communities, functions and functional genes (e.g., alkB, alkB1, XylE, NAH, PHE and BPH4). Biostimulation by the addition of SBNs proved an effective strategy for increasing the degradation of PHCs and facilitating the growth and metabolic activity of PHC degraders.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
9.60
自引率
10.40%
发文量
107
审稿时长
21 days
期刊介绍:
International Biodeterioration and Biodegradation publishes original research papers and reviews on the biological causes of deterioration or degradation.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


