鉴定降解氯化苯酚的最佳裂解酶:分子建模、对接、动力学和自由能计算的启示
Q1 Environmental Science
引用次数: 0
摘要
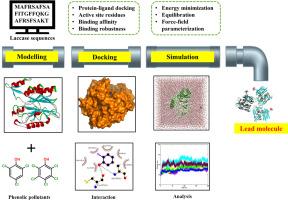
本研究旨在调查二十种不同的细菌和真菌漆酶与氯化酚(CPs)之间的相互作用,氯化酚被美国环境保护局(USEPA)列为优先污染物。为了节省时间和减少体外选择漆酶降解氯化石蜡的工作,研究人员利用分子对接技术从二十种漆酶中找出了降解氯化石蜡的最佳漆酶。氯化石蜡与细菌和真菌漆酶的平均结合能分别为- 4.72 kcal/mol和- 5.21 kcal/mol。根据最高的平均结合能,利用 200 毫微秒分子动力学模拟研究了多色曲霉(真菌)和 arboricola 黄单胞菌(细菌)漆酶的非结合型和结合型的稳定性和相互作用动力学。T. versicolor 和 X. arboricola 漆酶结合形式的均方根偏差(RMSD)分别在 0.25 nm 和 0.5 nm 的范围内。所有漆酶结合形式的回旋半径(Rg)保持在 1.88 至 2.24 nm 的范围内,大多数残基的均方根波动(RMSF)在 0.24 nm 以内,与漆酶的apo值一致。在水溶液中,在 300 K 和 1 bar 压力下,疏水相互作用、氢键和范德华力促进了漆酶与氯化石蜡的稳定性。相应的计算见解将有助于体外降解氯化石蜡的漆酶的选择、设计和工程应用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Identification of optimal laccases for chlorinated phenol degradation: Insights from molecular modelling, docking, dynamics, and free energy calculations
This study aimed to investigate the interactions between twenty different bacterial and fungal laccases and chlorinated phenols (CPs), which are listed as priority pollutants by the United States Environmental Protection Agency (USEPA). Molecular docking was used to identify the optimal laccase for degrading CPs out of twenty selected laccases to save time and reduce the in vitro selection of laccase for CPs degradation. The average binding energies of CPs with bacterial and fungal laccases were − 4.72 kcal/mol and − 5.21 kcal/mol, respectively. Based on the highest average binding energies Trametes versicolor (fungus) and Xanthomonas arboricola (bacteria) laccase apo and bound forms were investigated for stability and interaction dynamics using a 200 ns molecular dynamic simulation. The root mean square deviations (RMSD) of T. versicolor and X. arboricola laccase bound forms were within the 0.25 nm and 0.5 nm limit. The radius of gyration (Rg) for all laccases bound forms remained within the range from 1.88 to 2.24 nm with majority of residues in root mean square fluctuations (RMSF) were within the 0.24 nm and found consistent with the laccase apo values. In an aqueous solution, at ∼300 K and ∼ 1 bar pressure, laccase stability with CPs is facilitated by hydrophobic interactions, hydrogen bonds, and van der Waals forces. The corresponding computational insights will be advantageous for the selection, design and application in engineering of laccases for in vitro CPs degradation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Bioresource Technology Reports
Environmental Science-Environmental Engineering
CiteScore
7.20
自引率
0.00%
发文量
390
审稿时长
28 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


