健康人群咽部奈瑟菌的动态携带情况。
IF 2.6
4区 医学
Q3 INFECTIOUS DISEASES
引用次数: 0
摘要
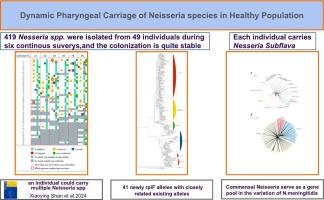
考虑到咽部携带的共生奈瑟菌对脑膜炎奈瑟菌的变异及其耐药基因的获得具有重要作用,了解人类咽部定植的奈瑟菌的真实种群具有重要意义。在这项研究中,我们对部分健康人群的口咽带菌情况进行了为期五个月的连续调查,以揭示不同奈瑟菌种的长期带菌情况。从 49 名参与者的 205 份咽拭子中的 203 份中分离出 419 株奈瑟菌。通过综合方法(MALDI-TOF-MS、rplF 测序和基因组测序),这些分离株被鉴定为亚弗拉伐奈瑟菌(N.subflava,n = 290)、粘膜奈瑟菌(N.mucosa,n = 52)、口腔奈瑟菌(N.oralis,n = 8)、细长奈瑟菌群(N.elongata,n = 6)和未确认菌种的奈瑟菌(n = 63)。所有个体和 168 份拭子(81.95%)中都分离出了亚扁桃体。从 25(45)、6(7)、4(5)和 20(53)个个体(拭子)中分别分离出粘膜奈瑟菌、口腔奈瑟菌、细长奈瑟菌和未确认菌种的奈瑟菌。从一个样本中分离出多个奈瑟氏菌属或一个菌种的多个克隆的情况很常见。在五个月内,一个人身上可能会频繁分离出相同的菌株。这些结果表明,奈瑟氏菌属和亚弗拉伐奈瑟氏菌在人的咽部无处不在,并且都有不同的种群;我们在研究脑膜炎奈瑟氏菌或其他呼吸道病原体时应更多地关注它们;鉴定奈瑟氏菌种的可靠而简便的方法仍有待开发。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Dynamic pharyngeal carriage of Neisseria species in healthy population
Considering the significant role of commensal Neisseria carried in the pharynx on the variation of N.meningitidis and the acquisition of its resistance genes, understanding the true Neisseria population colonizing the human pharynx is of great significance. In this study, we carried out a five-month continuous survey of oropharyngeal carriage in a certain healthy population to reveal the long-term carriage status of different Neisseria species. Totally, 419 Neisseria strains were isolated from 203 out of 205 pharyngeal swabs of 49 participants. Using combined methods (MALDI-TOF-MS, rplF sequencing and genome sequencing), the isolates were identified as N.subflava (n = 290), N.mucosa (n = 52), N.oralis (n = 8), N.elongata group (n = 6) and non-species-confirmed (n = 63). N.subflava was isolated from all individuals and 168 swabs (81.95 %). N.mucosa, N.oralis, N.elongata and non-species-confirmed were isolated from 25 (45), 6 (7), 4 (5) and 20 (53) individuals (swabs) respectively. It was common that multiple Neisseria spp. or multiple clones of one species were isolated from a single sample. An identical strain could be isolated frequently from a single person within five months. These results indicate that Neisseria spp. and N.subflava are ubiquitous in human pharynx and both have diverse population; we should pay more attention to them when studying N.meningitidis or other respiratory pathogens; robust and handy method for identifying Neisseria species remains to be developed.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Infection Genetics and Evolution
医学-传染病学
CiteScore
8.40
自引率
0.00%
发文量
215
审稿时长
82 days
期刊介绍:
(aka Journal of Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases -- MEEGID)
Infectious diseases constitute one of the main challenges to medical science in the coming century. The impressive development of molecular megatechnologies and of bioinformatics have greatly increased our knowledge of the evolution, transmission and pathogenicity of infectious diseases. Research has shown that host susceptibility to many infectious diseases has a genetic basis. Furthermore, much is now known on the molecular epidemiology, evolution and virulence of pathogenic agents, as well as their resistance to drugs, vaccines, and antibiotics. Equally, research on the genetics of disease vectors has greatly improved our understanding of their systematics, has increased our capacity to identify target populations for control or intervention, and has provided detailed information on the mechanisms of insecticide resistance.
However, the genetics and evolutionary biology of hosts, pathogens and vectors have tended to develop as three separate fields of research. This artificial compartmentalisation is of concern due to our growing appreciation of the strong co-evolutionary interactions among hosts, pathogens and vectors.
Infection, Genetics and Evolution and its companion congress [MEEGID](http://www.meegidconference.com/) (for Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases) are the main forum acting for the cross-fertilization between evolutionary science and biomedical research on infectious diseases.
Infection, Genetics and Evolution is the only journal that welcomes articles dealing with the genetics and evolutionary biology of hosts, pathogens and vectors, and coevolution processes among them in relation to infection and disease manifestation. All infectious models enter the scope of the journal, including pathogens of humans, animals and plants, either parasites, fungi, bacteria, viruses or prions. The journal welcomes articles dealing with genetics, population genetics, genomics, postgenomics, gene expression, evolutionary biology, population dynamics, mathematical modeling and bioinformatics. We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services .
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


