在气道感染过程中,铜绿假单胞菌面临着粘膜定植和抗生素耐受性之间的适应性权衡
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
铜绿假单胞菌经常引起抗生素拮抗性肺炎,但其在人类感染过程中的适应机制尚不清楚。为了揭示铜绿假单胞菌在粘膜表面的选择性压力和适应策略,我们通过转座子插入测序(Tn-seq)研究了铜绿假单胞菌在组织工程气道中的生长和抗生素耐受性。基于 Tn-seq 数据的代谢模型揭示了铜绿假单胞菌生长的营养需求,突出了对葡萄糖和乳酸的依赖以及对氨基酸生物合成的不同需求。Tn-seq 还揭示了在没有抗生素的情况下粘膜生长过程中对生物膜形成的选择。工程有机体的实时成像显示,生物膜栖息细胞在粘膜表面定殖时保持无柄状态,从而限制了营养物质的觅食并降低了生长速度。相反,生物膜的形成增加了粘膜表面对抗生素的耐受性。此外,生物膜表型加剧的突变体保护了耐受性较差但细胞毒性更强的菌株,从而促进了表型的异质性。因此,铜绿假单胞菌必须在相互冲突的物理和生物选择压力下建立慢性感染。本文章由计算机程序翻译,如有差异,请以英文原文为准。
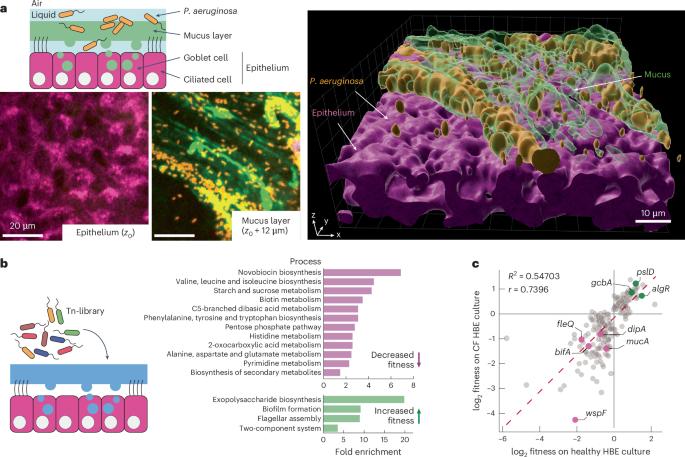
Pseudomonas aeruginosa faces a fitness trade-off between mucosal colonization and antibiotic tolerance during airway infection
Pseudomonas aeruginosa frequently causes antibiotic-recalcitrant pneumonia, but the mechanisms driving its adaptation during human infections remain unclear. To reveal the selective pressures and adaptation strategies at the mucosal surface, here we investigated P. aeruginosa growth and antibiotic tolerance in tissue-engineered airways by transposon insertion sequencing (Tn-seq). Metabolic modelling based on Tn-seq data revealed the nutritional requirements for P. aeruginosa growth, highlighting reliance on glucose and lactate and varying requirements for amino acid biosynthesis. Tn-seq also revealed selection against biofilm formation during mucosal growth in the absence of antibiotics. Live imaging in engineered organoids showed that biofilm-dwelling cells remained sessile while colonizing the mucosal surface, limiting nutrient foraging and reduced growth. Conversely, biofilm formation increased antibiotic tolerance at the mucosal surface. Moreover, mutants with exacerbated biofilm phenotypes protected less tolerant but more cytotoxic strains, contributing to phenotypic heterogeneity. P. aeruginosa must therefore navigate conflicting physical and biological selective pressures to establish chronic infections. Tn-seq and live imaging of Pseudomonas aeruginosa during airway epithelial organoid infections reveal metabolic adaptations and trade-offs between growth and antibiotic tolerance in biofilm lifestyle, impacting bacterial spread and cytotoxicity.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


