从稀疏且有噪声的单分子 X 射线散射图像中确定贝叶斯电子密度
IF 11.7
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
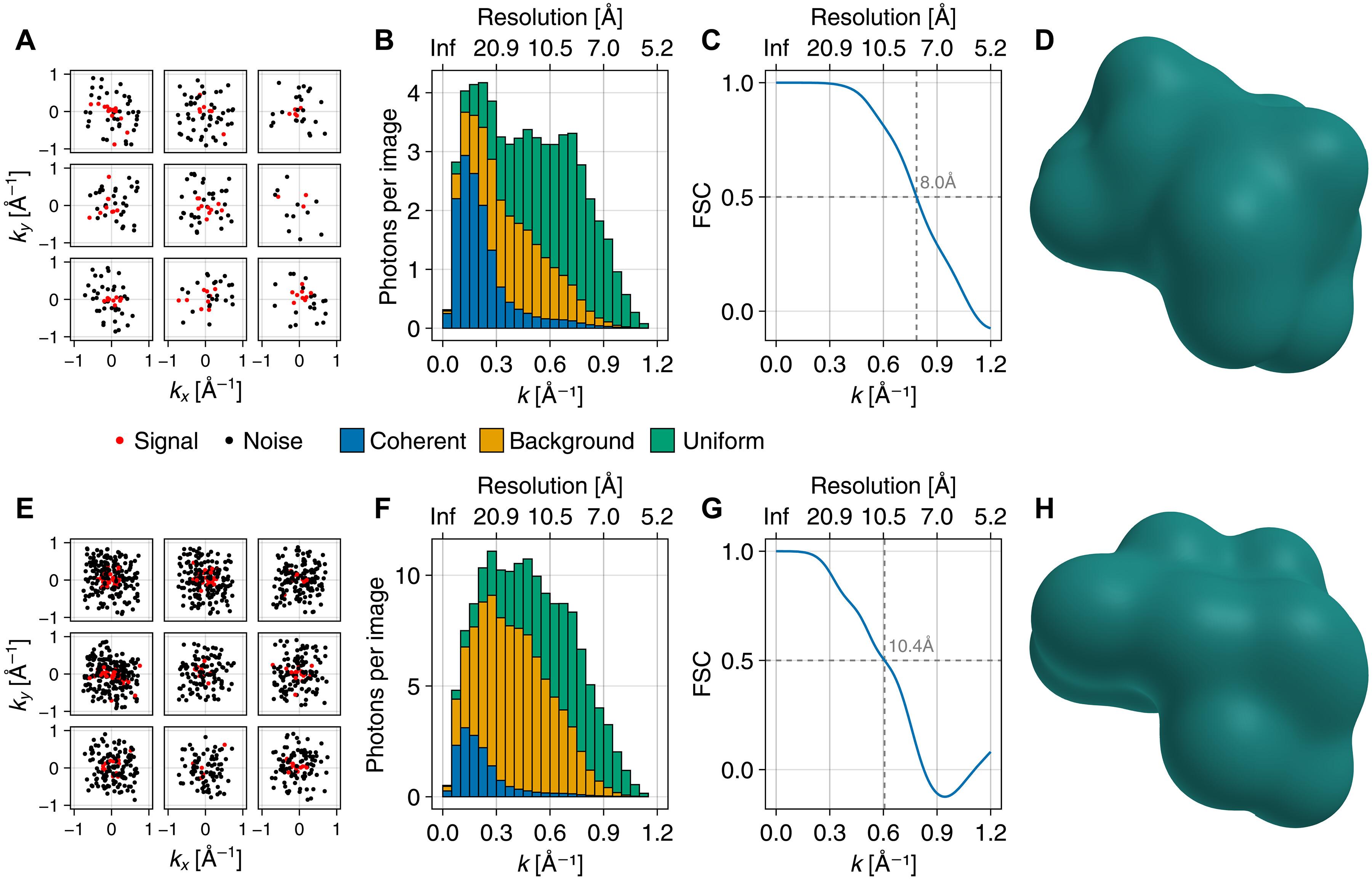
使用自由电子激光器进行的单分子 X 射线散射实验具有解析生物分子结构和结构组合的潜力。然而,由于光子数少、噪声大、命中率低,迄今为止还无法实现分子电子密度的测定。因此,大多数方法都侧重于大型标本(如整个病毒),这些标本会散射足够多的光子,从而可以确定每幅图像的方向。而蛋白质等小样本散射的光子太少,无法确定分子方向。在这里,我们提出了一种严格的贝叶斯方法来克服这些局限性,此外还考虑了强度波动、光束偏振、不规则探测器形状、非相干散射和背景散射。我们利用合成散射图像证明,在这种极端高噪声泊松机制下,可以测定小蛋白质的电子密度。对已公布的病毒数据进行的测试达到了 9 nm 的探测器极限分辨率,每幅图像只使用了可用光子的 0.01%。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Bayesian electron density determination from sparse and noisy single-molecule X-ray scattering images
Single molecule x-ray scattering experiments using free-electron lasers hold the potential to resolve biomolecular structures and structural ensembles. However, molecular electron density determination has so far not been achieved because of low photon counts, high noise levels, and low hit rates. Most approaches therefore focus on large specimen like entire viruses, which scatter sufficiently many photons to allow orientation determination of each image. Small specimens like proteins, however, scatter too few photons for the molecular orientations to be determined. Here, we present a rigorous Bayesian approach to overcome these limitations, additionally taking into account intensity fluctuations, beam polarization, irregular detector shapes, incoherent scattering, and background scattering. We demonstrate using synthetic scattering images that electron density determination of small proteins is possible in this extreme high noise Poisson regime. Tests on published virus data achieved the detector-limited resolution of 9 nm, using only 0.01% of the available photons per image.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


