与 DPANN 古菌超门相关的病毒和移动遗传元素的元基因组特征
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
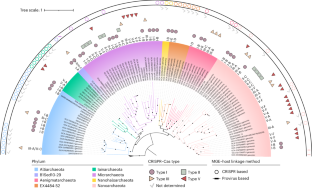
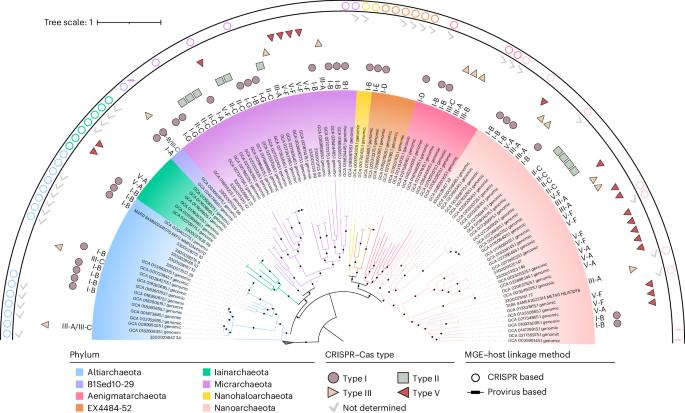
古菌超门 DPANN(由最先发现的五个门的首字母缩写组成:DPANN是一个超小型共生菌群,能够在极端生态系统中生存。DPANN 古菌及其病毒体之间的多样性和动态关系在很大程度上仍不为人所知。在这里,我们使用一种元基因组聚类规律性间隔短回文重复序列(CRISPR)筛选方法,鉴定出 97 种全球分布的非冗余病毒和未分类的移动遗传因子,预测它们会感染 8 个 DPANN 门的宿主,其中包括 7 个以前没有表征过的病毒群。基因组分析表明了病毒形态的多样性,包括有头尾病毒、无尾二十面体病毒和纺锤形病毒,它们都有可能造成溶解性、慢性或溶源性感染。我们还发现了病毒编码的 Cas12f1 蛋白(可能源自未培养的 DPANN 古菌)和微型CRISPR 阵列的证据,它们可能在调节宿主新陈代谢方面发挥作用。许多元基因组的病毒与宿主比率为10,这表明DPANN病毒在控制宿主种群方面发挥着重要作用。总之,我们的研究揭示了 DPANN 病毒组未被充分探索的多样性、功能谱系和宿主相互作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Metagenomic characterization of viruses and mobile genetic elements associated with the DPANN archaeal superphylum
The archaeal superphylum DPANN (an acronym formed from the initials of the first five phyla discovered: Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanohaloarchaeota and Nanoarchaeota) is a group of ultrasmall symbionts able to survive in extreme ecosystems. The diversity and dynamics between DPANN archaea and their virome remain largely unknown. Here we use a metagenomic clustered regularly interspaced short palindromic repeats (CRISPR) screening approach to identify 97 globally distributed, non-redundant viruses and unclassified mobile genetic elements predicted to infect hosts across 8 DPANN phyla, including 7 viral groups not previously characterized. Genomic analysis suggests a diversity of viral morphologies including head-tailed, tailless icosahedral and spindle-shaped viruses with the potential to establish lytic, chronic or lysogenic infections. We also find evidence of a virally encoded Cas12f1 protein (probably originating from uncultured DPANN archaea) and a mini-CRISPR array, which could play a role in modulating host metabolism. Many metagenomes have virus-to-host ratios >10, indicating that DPANN viruses play an important role in controlling host populations. Overall, our study illuminates the underexplored diversity, functional repertoires and host interactions of the DPANN virome. A metagenomic CRISPR screening approach identifies an expansive diversity of viruses and mobile genetic elements infecting the enigmatic ultrasmall DPANN archaea.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


