RNA 基础结构剖析揭示拥挤空间中的转录组结构
IF 6.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
RNA 在细胞中折叠成紧凑的结构,并与蛋白质发生相互作用。这些闭塞的环境会阻挡探测底层 RNA 的试剂。能在拥挤环境中分析结构的探针可以揭示 RNA 的生物学特性。在这里,我们采用的 2′-OH 反应探针足够小,可以进入细胞内分子接触紧密的底层折叠 RNA 结构,为细胞内 RNA 结构分析提供更广泛的覆盖范围。这些数据首先通过特性良好的人类核糖体 RNA 进行分析,然后应用于整个转录组的多聚腺苷酸转录本。最小的探针乙酰咪唑(AcIm)的结构覆盖率比较大的传统试剂 NAIN3 高出 80%,为数百个转录本提供了更多的结构信息。乙酰探针还能提供识别转录本中 m6A 修饰位点的卓越信号,尤其是在标准探针无法到达的位点。我们的策略能够剖析 RNA 基础结构,加强对转录本组结构、修饰和细胞内相互作用的分析,尤其是在空间拥挤的环境中。本文章由计算机程序翻译,如有差异,请以英文原文为准。
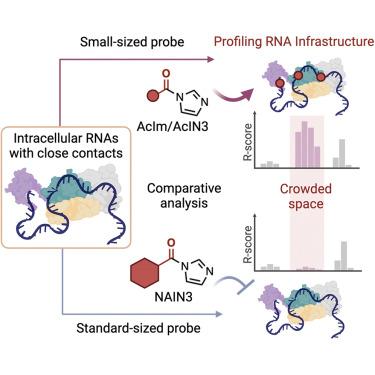

RNA infrastructure profiling illuminates transcriptome structure in crowded spaces
RNAs fold into compact structures and undergo protein interactions in cells. These occluded environments can block reagents that probe the underlying RNAs. Probes that can analyze structure in crowded settings can shed light on RNA biology. Here, we employ 2′-OH-reactive probes that are small enough to access folded RNA structure underlying close molecular contacts within cells, providing considerably broader coverage for intracellular RNA structural analysis. The data are analyzed first with well-characterized human ribosomal RNAs and then applied transcriptome-wide to polyadenylated transcripts. The smallest probe acetylimidazole (AcIm) yields 80% greater structural coverage than larger conventional reagent NAIN3, providing enhanced structural information in hundreds of transcripts. The acetyl probe also provides superior signals for identifying m6A modification sites in transcripts, particularly in sites that are inaccessible to a standard probe. Our strategy enables profiling RNA infrastructure, enhancing analysis of transcriptome structure, modification, and intracellular interactions, especially in spatially crowded settings.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Chemical Biology
Biochemistry, Genetics and Molecular Biology-Molecular Medicine
CiteScore
14.70
自引率
2.30%
发文量
143
期刊介绍:
Cell Chemical Biology, a Cell Press journal established in 1994 as Chemistry & Biology, focuses on publishing crucial advances in chemical biology research with broad appeal to our diverse community, spanning basic scientists to clinicians. Pioneering investigations at the chemistry-biology interface, the journal fosters collaboration between these disciplines. We encourage submissions providing significant conceptual advancements of broad interest across chemical, biological, clinical, and related fields. Particularly sought are articles utilizing chemical tools to perturb, visualize, and measure biological systems, offering unique insights into molecular mechanisms, disease biology, and therapeutics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


