利用 LABEL-seq 对细胞内蛋白质的丰度、活性、相互作用和可药性进行多重分析。
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
在这里,我们介绍了用条形码标记和富集测序进行生化分析(LABEL-seq),这是一种大规模并行分析人体细胞中集合蛋白质变体的方法。通过利用 RNA 结合域(RBD)与稳定的变体编码 RNA 条形码在细胞内的自组装,LABEL-seq 利用 RBD 蛋白融合体的简单亲和富集,然后对共同富集的条形码进行高通量测序,促进了蛋白质特性和功能的直接测量。对约 1,600 个 BRaf 变体的约 20,000 种变异效应进行测量后发现,癌症中经常发生变异的位置的变异对细胞内丰度的影响微乎其微,但却能显著改变活性、蛋白质间相互作用和可药性。综合分析确定了具有类似生化作用的位置网络,并建立了变体对细胞增殖和小分子促进降解的影响模型。因此,LABEL-seq 能够在原生细胞环境中直接测量多种生化特性,从而深入了解蛋白质功能、疾病机制和可药性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
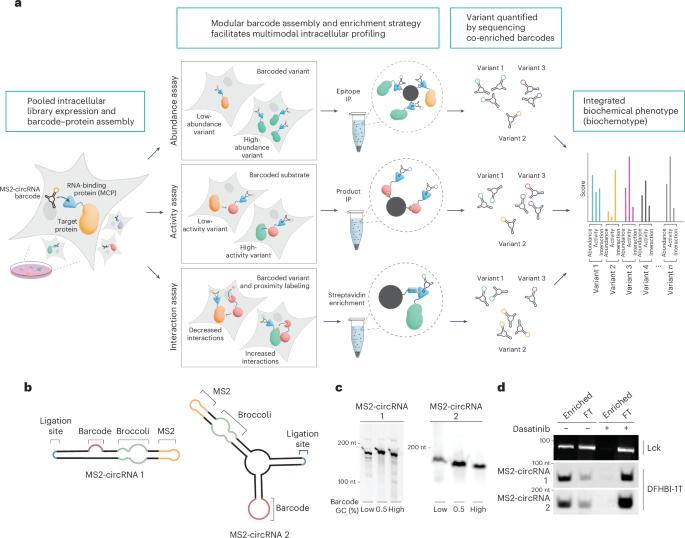
Multiplexed profiling of intracellular protein abundance, activity, interactions and druggability with LABEL-seq
Here we describe labeling with barcodes and enrichment for biochemical analysis by sequencing (LABEL-seq), an assay for massively parallel profiling of pooled protein variants in human cells. By leveraging the intracellular self-assembly of an RNA-binding domain (RBD) with a stable, variant-encoding RNA barcode, LABEL-seq facilitates the direct measurement of protein properties and functions using simple affinity enrichments of RBD protein fusions, followed by high-throughput sequencing of co-enriched barcodes. Measurement of ~20,000 variant effects for ~1,600 BRaf variants revealed that variation at positions frequently mutated in cancer minimally impacted intracellular abundance but could dramatically alter activity, protein–protein interactions and druggability. Integrative analysis identified networks of positions with similar biochemical roles and enabled modeling of variant effects on cell proliferation and small molecule-promoted degradation. Thus, LABEL-seq enables direct measurement of multiple biochemical properties in a native cellular context, providing insights into protein function, disease mechanisms and druggability. Labeling with barcodes and enrichment for biochemical analysis by sequencing (LABEL-seq) enables massively parallel profiling of thousands of pooled protein variants in cells, yielding insight into protein function, interactions and druggability.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


