构建泛基因组图谱
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
泛基因组图谱可以代表多个参考基因组之间的所有变异,但目前构建泛基因组图谱的方法排除了复杂序列,或者基于单一参考。为此,我们开发了泛基因组图谱生成器(PanGenome Graph Builder),这是一种构建泛基因组图谱的流水线,不会产生偏差或排斥。PanGenome Graph Builder 使用全对全比对来构建变异图,我们可以在其中识别变异、测量保护、检测重组事件并推断系统发育关系。本文章由计算机程序翻译,如有差异,请以英文原文为准。
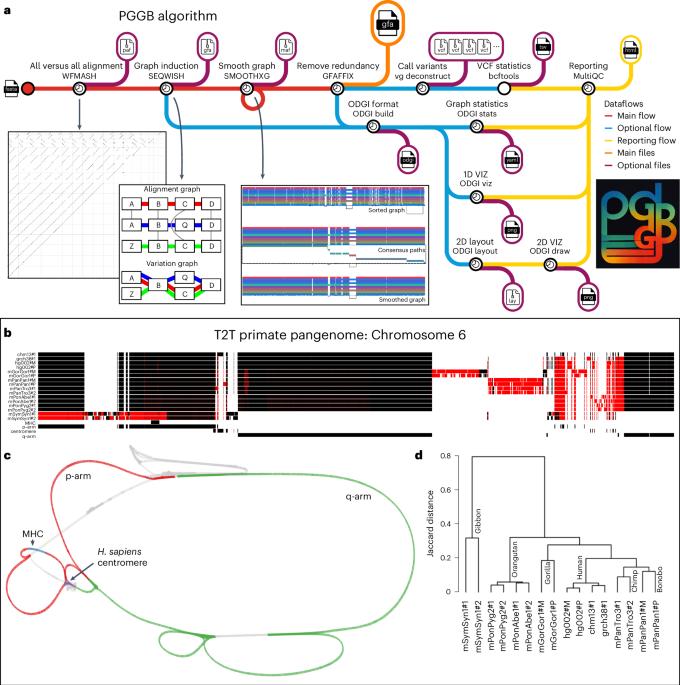
Building pangenome graphs
Pangenome graphs can represent all variation between multiple reference genomes, but current approaches to build them exclude complex sequences or are based upon a single reference. In response, we developed the PanGenome Graph Builder, a pipeline for constructing pangenome graphs without bias or exclusion. The PanGenome Graph Builder uses all-to-all alignments to build a variation graph in which we can identify variation, measure conservation, detect recombination events and infer phylogenetic relationships. PGGB is a modular framework for efficiently building unbiased pangenome graphs, supporting diverse downstream analyses.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


