基于人工智能的高精度热葡聚糖地衣芽孢杆菌酶约束模型的自动构建。
IF 6.8
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
Geobacillus thermoglucosidasius NCIMB 11955 具有耐高温、生长速度快、污染风险低等优点。此外,它还具有高效的基因编辑工具,是最有前途的下一代细胞工厂之一。然而,作为一种非模型微生物,代谢信息的缺乏极大地阻碍了高精度代谢通量模型的构建。在此,我们提出了一种基于人工智能技术的生物智能模型(BIM)策略,用于自动构建酶约束模型。1) .BIM 利用对比学习酶注释(CLEAN)预测工具来分析热葡糖球菌 NCIMB 11955 的整个基因组序列,从而发现非模型菌株中潜在的功能蛋白。2).BIM 的 MetaPatchM 模块可自动修复代谢网络模型。3).天津科技大学-kcat(TUST-kcat)模块可预测模型中酶的 kcat 值。4).4). Enzyme-insert 程序构建酶约束模型,并进行全局扫描以解决过度约束问题。酶数据被自动整合到代谢通量模型中,创建了一个酶约束模型 ec_G-ther11955。为了验证模型的准确性,我们使用 p-thermo 和 ec_G-ther11955 模型预测核黄素生产策略。ec_G-ther11955模型的准确性明显更高。为了进一步验证其有效性,我们使用 ec_G-ther11955 来指导合理设计生产 L-缬氨酸的菌株。利用识别导致定向过量生产的所有遗传操作的优化程序(OptForce)、预测性敲除靶标(PKT)和基于强制目标通量的通量扫描(FSEOF)算法,我们识别出了 24 个敲除和过表达靶标,准确率达到 87.5%。最终,L-缬氨酸滴度提高了 664.04%。这项研究为快速构建非模型菌株模型提供了一种新策略,并展示了人工智能在代谢工程中的巨大潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
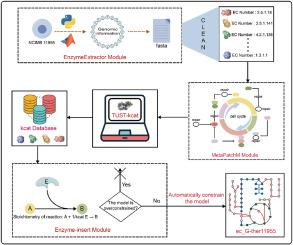
AI-based automated construction of high-precision Geobacillus thermoglucosidasius enzyme constraint model
Geobacillus thermoglucosidasius NCIMB 11955 possesses advantages, such as high-temperature tolerance, rapid growth rate, and low contamination risk. Additionally, it features efficient gene editing tools, making it one of the most promising next-generation cell factories. However, as a non-model microorganism, a lack of metabolic information significantly hampers the construction of high-precision metabolic flux models. Here, we propose a BioIntelliModel (BIM) strategy based on artificial intelligence technology for the automated construction of enzyme-constrained models. 1). BIM utilises the Contrastive Learning Enabled Enzyme Annotation (CLEAN) prediction tool to analyse the entire genome sequence of G. thermoglucosidasius NCIMB 11955, uncovering potential functional proteins in non-model strains. 2). The MetaPatchM module of BIM automates the repair of the metabolic network model. 3). The Tianjin University of Science and Technology-kcat (TUST-kcat) module predicts the kcat values of enzymes within the model. 4). The Enzyme-insert procedure constructs an enzyme-constrained model and performs a global scan to address overconstraint issues. Enzymatic data were automatically integrated into the metabolic flux model, creating an enzyme-constrained model, ec_G-ther11955. To validate model accuracy, we used both the p-thermo and ec_G-ther11955 models to predict riboflavin production strategies. The ec_G-ther11955 model demonstrated significantly higher accuracy. To further verify its efficacy, we employed ec_G-ther11955 to guide the rational design of L-valine-producing strains. Using the Optimisation Procedure for Identifying All Genetic Manipulations Leading to Targeted Overproductions (OptForce), Predictive Knockout Targeting (PKT), and Flux Scanning based on Enforced Objective Flux (FSEOF) algorithms, we identified 24 knockout and overexpression targets, achieving an accuracy rate of 87.5%. Ultimately, this led to an increase of 664.04% in L-valine titre. This study provides a novel strategy for rapidly constructing non-model strain models and demonstrates the tremendous potential of artificial intelligence in metabolic engineering.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Metabolic engineering
工程技术-生物工程与应用微生物
CiteScore
15.60
自引率
6.00%
发文量
140
审稿时长
44 days
期刊介绍:
Metabolic Engineering (MBE) is a journal that focuses on publishing original research papers on the directed modulation of metabolic pathways for metabolite overproduction or the enhancement of cellular properties. It welcomes papers that describe the engineering of native pathways and the synthesis of heterologous pathways to convert microorganisms into microbial cell factories. The journal covers experimental, computational, and modeling approaches for understanding metabolic pathways and manipulating them through genetic, media, or environmental means. Effective exploration of metabolic pathways necessitates the use of molecular biology and biochemistry methods, as well as engineering techniques for modeling and data analysis. MBE serves as a platform for interdisciplinary research in fields such as biochemistry, molecular biology, applied microbiology, cellular physiology, cellular nutrition in health and disease, and biochemical engineering. The journal publishes various types of papers, including original research papers and review papers. It is indexed and abstracted in databases such as Scopus, Embase, EMBiology, Current Contents - Life Sciences and Clinical Medicine, Science Citation Index, PubMed/Medline, CAS and Biotechnology Citation Index.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


