通过将临床表型与可量化的免疫组群成分联系起来,实现可解释的基因组学分析。
IF 5.1
1区 生物学
Q1 BIOLOGY
引用次数: 0
摘要
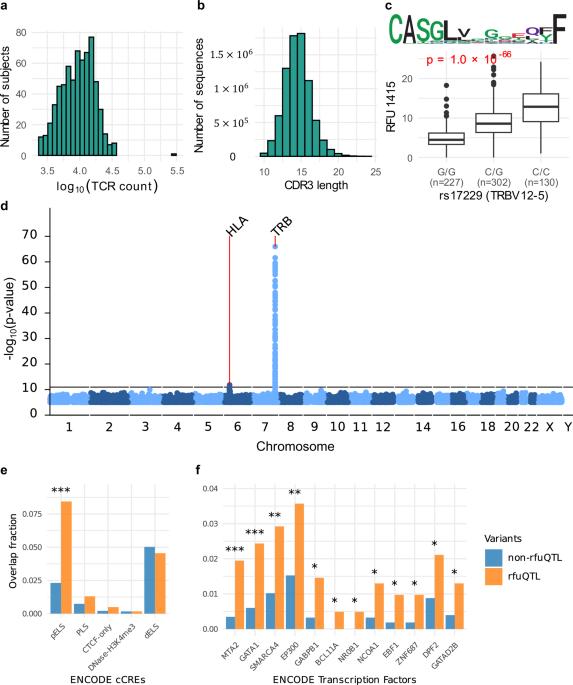
在 GWAS 研究中弥合基因型与表型之间的差距具有挑战性。许多基因变异与包括癌症在内的免疫相关疾病有关,但大多数变异的可解释性仍然很低。在这里,我们研究了 T 细胞受体(TCR)复合物中的定量成分,即预测具有共同抗原特异性的 TCR 序列群的频率,以解释人类各种疾病的遗传关联。我们首先开发了一个统计模型,利用 TRB 和 HLA 基因座的变异来预测 TCR 成分。将该模型应用于英国生物库数据中的 30 多万人,我们发现了 2309 种 TCR 丰度与各种免疫疾病之间的关联。预测对自身免疫性疾病具有致病性的 TCR 簇明显富集于预测的自身抗原特异性。此外,有四个 TCR 簇与不同癌症的较佳预后相关,而传统的 GWAS 无法在这些癌症中发现任何重要的基因位点。总之,我们的研究结果凸显了适应性免疫反应在解释基因型与表型之间的关联方面所起的不可或缺的作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Interpretable GWAS by linking clinical phenotypes to quantifiable immune repertoire components
Bridging the gap between genotype and phenotype in GWAS studies is challenging. A multitude of genetic variants have been associated with immune-related diseases, including cancer, yet the interpretability of most variants remains low. Here, we investigate the quantitative components in the T cell receptor (TCR) repertoire, the frequency of clusters of TCR sequences predicted to have common antigen specificity, to interpret the genetic associations of diverse human diseases. We first developed a statistical model to predict the TCR components using variants in the TRB and HLA loci. Applying this model to over 300,000 individuals in the UK Biobank data, we identified 2309 associations between TCR abundances and various immune diseases. TCR clusters predicted to be pathogenic for autoimmune diseases were significantly enriched for predicted autoantigen-specificity. Moreover, four TCR clusters were associated with better outcomes in distinct cancers, where conventional GWAS cannot identify any significant locus. Collectively, our results highlight the integral role of adaptive immune responses in explaining the associations between genotype and phenotype. Analyzing the impact of genetic variants on T cell receptor repertoire components reveals the mechanisms behind susceptibility variants in autoimmune diseases and cancers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


