对顺铂诱导的 A549 细胞凋亡进行全局蛋白质组学和磷酸蛋白质组学综合分析。
IF 2.5
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
Biochemical and biophysical research communications
Pub Date : 2024-10-18
DOI:10.1016/j.bbrc.2024.150846
引用次数: 0
摘要
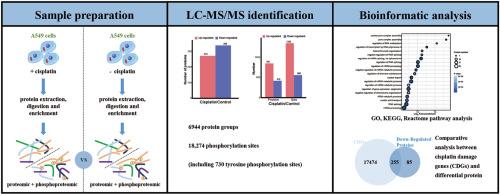
蛋白质磷酸化是一种广泛存在的重要翻译后修饰,与各种生物过程密不可分。我们以前曾利用蛋白质亲和探针来识别顺铂损伤的基因,结果发现顺铂对蛋白激酶和蛋白磷酸酶基因造成了严重损伤。在本研究中,我们研究了顺铂诱导的 A549 细胞全蛋白质组和磷酸化蛋白质组的改变。利用Fe-IMAC珠和酪氨酸磷酸化富集抗体,我们在顺铂处理的A549细胞和对照细胞的三个生物重复中鉴定出了6944个蛋白质组和4915个蛋白质上的18274个磷酸化位点。其中,发现了 730 个酪氨酸磷酸化位点,这是顺铂处理后在 A549 细胞中发现此类位点最多的一次。生物信息学分析表明,磷酸化水平发生显著变化的蛋白质主要参与 RNA 处理、修饰、转录、翻译和剪接体。这表明,顺铂诱导的蛋白激酶和磷酸酶损伤可能会破坏这些蛋白的正常功能,从而影响 DNA 复制、RNA 翻译和剪切,最终导致肿瘤细胞死亡。此外,我们还将蛋白质组数据与之前获得的顺铂损伤基因进行了交叉比对,发现大部分下调的蛋白质都来自于顺铂诱导的基因损伤。这些数据可在 ProteomeXchange 上查阅,其标识符为 PXD053902。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Integrated global proteomic and phosphoproteomic analysis of cisplatin-induced apoptosis in A549 cells
Protein phosphorylation, a widely occurring and significant post-translational modification, is integral to various biological processes. We previously utilized a protein affinity probe to identify genes damaged by cisplatin, revealing that it inflicts substantial damage on protein kinase and protein phosphatase genes. In this study, we investigated cisplatin-induced alterations in the global proteome and phosphoproteome of A549 cells. Employing Fe-IMAC beads and tyrosine phosphorylation enrichment antibodies, we identified 6944 protein groups and 18,274 phosphorylation sites on 4915 proteins across three biological replicates of both cisplatin-treated A549 cells and control cells. Among these, 730 tyrosine phosphorylation sites were identified—marking the most substantial discovery of such sites in A549 cells following cisplatin treatment. Bioinformatics analysis indicated that the proteins exhibiting significant phosphorylation level changes predominantly involved in RNA processing, modification, transcription, translation, and the spliceosome. This suggests that cisplatin-induced damage to protein kinases and phosphatases may disrupt the normal function of these proteins, consequently impairing DNA replication, RNA translation, and shearing, ultimately culminating in tumor cell death. Moreover, we cross-referenced our proteomic data with our previously obtained cisplatin-damaged genes, observing that the majority of down-regulated proteins derived from cisplatin-induced gene damage. The data are available on ProteomeXchange under the identifier PXD053902.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
6.10
自引率
0.00%
发文量
1400
审稿时长
14 days
期刊介绍:
Biochemical and Biophysical Research Communications is the premier international journal devoted to the very rapid dissemination of timely and significant experimental results in diverse fields of biological research. The development of the "Breakthroughs and Views" section brings the minireview format to the journal, and issues often contain collections of special interest manuscripts. BBRC is published weekly (52 issues/year).Research Areas now include: Biochemistry; biophysics; cell biology; developmental biology; immunology
; molecular biology; neurobiology; plant biology and proteomics

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


