深度挖掘揭示脊椎动物基因组中内源性病毒元件的多样性
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
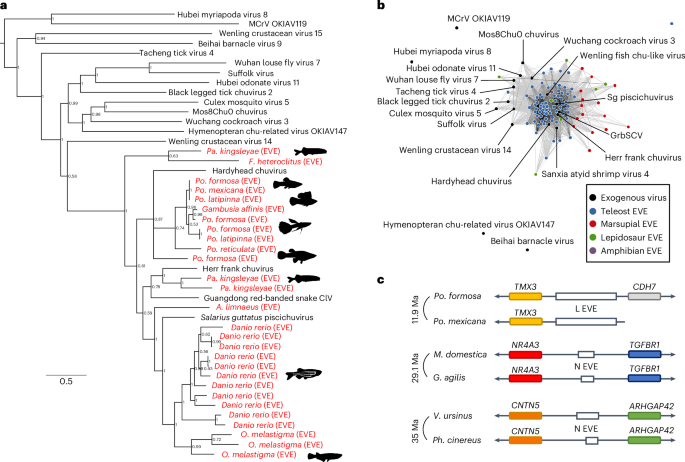
病毒与宿主基因组的整合会产生内源性病毒元件(EVEs),从而为了解病毒的多样性、宿主范围和进化提供了线索。考虑到现有的基因组数据,系统地搜索 EVEs 在计算上越来越具有挑战性。我们使用云计算方法对脊椎动物中的Shotokuvirae和Orthornavirae进行了全面的EVEs搜索。我们在 295 个脊椎动物基因组中发现了 2,040 个 EVEs,并提供了属于 Chuviridae、Paramyxoviridae、Nairoviridae 和 Benyviridae 科的 EVEs 的证据。我们还发现了一种来自黄病毒属的 EVE,它与鼠类啮齿动物具有同源关系。此外,我们的分析还发现,逆转录病毒和丝状病毒可能三次独立地从逆转录病毒元件中获得了它们的糖蛋白外结构域。总之,这些发现促使脊椎动物的病毒化石记录中增加了4个病毒科和肝病毒属,为了解它们的自然史和进化史提供了重要线索。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Deep mining reveals the diversity of endogenous viral elements in vertebrate genomes
Integration of viruses into host genomes can give rise to endogenous viral elements (EVEs), which provide insights into viral diversity, host range and evolution. A systematic search for EVEs is becoming computationally challenging given the available genomic data. We used a cloud-computing approach to perform a comprehensive search for EVEs in the kingdoms Shotokuvirae and Orthornavirae across vertebrates. We identified 2,040 EVEs in 295 vertebrate genomes and provide evidence for EVEs belonging to the families Chuviridae, Paramyxoviridae, Nairoviridae and Benyviridae. We also find an EVE from the Hepacivirus genus of flaviviruses with orthology across murine rodents. In addition, our analyses revealed that reptarenaviruses and filoviruses probably acquired their glycoprotein ectodomains three times independently from retroviral elements. Taken together, these findings encourage the addition of 4 virus families and the Hepacivirus genus to the growing virus fossil record of vertebrates, providing key insights into their natural history and evolution. Computational cloud-based screen of vertebrate genomes identifies endogenous viral elements of members of the kingdoms Shotokuvirae and Orthornavirae and informs about evolutionary history of non-retroviral elements.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


