用柔性酶重编遗传密码
IF 51.7
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
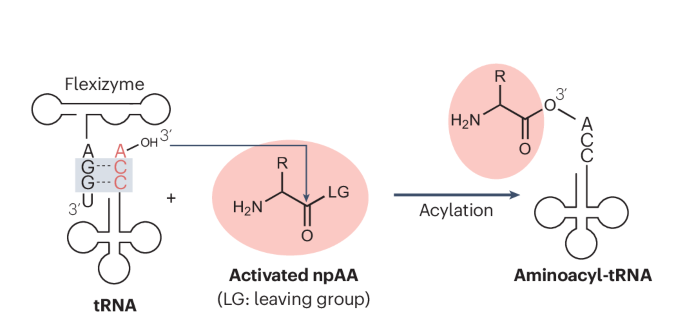
在规范遗传密码中,61 个有义密码子被分配给 20 个蛋白质氨基酸。随着遗传密码操作技术的进步,核糖体中可以加入非蛋白源氨基酸(npAAs)。将信使核糖核酸(mRNA)翻译成肽序列的关键分子是氨基酰转移核糖核酸(tRNA),它通过反密码子识别 mRNA 密码子。由于氨基酰-tRNA 合成酶(ARSs)对各自的氨基酸-tRNA 对具有高度特异性,因此使用天然 ARSs 制备 npAA-tRNAs 是不可行的。然而,柔性酶是一种适应性很强的氨基酰化核糖酶,可以利用被适当离去基团激活的氨基酸随意制备各种氨基酰-tRNA。在识别元件方面,柔性酶只需要活化氨基酸的离去基团或侧链中的芳香环,以及 tRNA 的保守 3′ 端 CCA。因此,柔性酶几乎可以将任何氨基酸加载到任何 tRNA 上。柔性酶系统不仅能处理侧链修饰的 l-α 氨基酸,还能处理各种骨架修饰的 npAA。本综述介绍了柔性酶变体的发展,讨论了它们的结构和机理,以及它们在遗传密码重编程合成独特多肽和蛋白质中的应用。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Reprogramming the genetic code with flexizymes
In the canonical genetic code, the 61 sense codons are assigned to the 20 proteinogenic amino acids. Advancements in genetic code manipulation techniques have enabled the ribosomal incorporation of nonproteinogenic amino acids (npAAs). The critical molecule for translating messenger RNA (mRNA) into peptide sequences is aminoacyl-transfer RNA (tRNA), which recognizes the mRNA codon through its anticodon. Because aminoacyl-tRNA synthetases (ARSs) are highly specific for their respective amino acid–tRNA pairs, it is not feasible to use natural ARSs to prepare npAA-tRNAs. However, flexizymes are adaptable aminoacylation ribozymes that can be used to prepare diverse aminoacyl-tRNAs at will using amino acids activated with suitable leaving groups. Regarding recognition elements, flexizymes require only an aromatic ring in either the leaving group or side chain of the activated amino acid, and the conserved 3′-end CCA of the tRNA. Therefore, flexizymes allow virtually any amino acid to be charged onto any tRNA. The flexizyme system can handle not only l-α-amino acids with side chain modifications but also various backbone-modified npAAs. This Review describes the development of flexizyme variants and discusses their structure and mechanism and their applications in genetic code reprogramming for the synthesis of unique peptides and proteins. Flexizymes can be used to prepare nonproteinogenic aminoacyl-tRNAs (npAA-tRNAs) by activating amino acids with suitable leaving groups, leading to ribosomal incorporation of npAAs by genetic code reprogramming. This enables the ribosomal synthesis of unique peptides and proteins containing various npAAs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature reviews. Chemistry
Chemical Engineering-General Chemical Engineering
CiteScore
52.80
自引率
0.80%
发文量
88
期刊介绍:
Nature Reviews Chemistry is an online-only journal that publishes Reviews, Perspectives, and Comments on various disciplines within chemistry. The Reviews aim to offer balanced and objective analyses of selected topics, providing clear descriptions of relevant scientific literature. The content is designed to be accessible to recent graduates in any chemistry-related discipline while also offering insights for principal investigators and industry-based research scientists. Additionally, Reviews should provide the authors' perspectives on future directions and opinions regarding the major challenges faced by researchers in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


