表观基因组异质性是肿瘤演变的源头
IF 72.5
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
在过去的十年中,通过开发针对 DNA 序列变异的治疗方法,癌症医学取得了显著进展。然而,由于同一基因型可产生不同的细胞状态,因此纯遗传的治疗选择方法受到了阻碍。在多细胞生物体中,细胞状态的异质性是由调控 DNA 功能(如转录)的表观遗传过程驱动的;这些过程的破坏是癌症的一个特征,它使有缺陷的细胞状态得以出现。单细胞技术的进步使我们有能力量化肿瘤的表观基因组异质性并了解其机制,从而改变我们对表观基因组变化如何驱动癌症进化的认识。本综述探讨了表观基因组异质性和可塑性是细胞状态的储存库,因此也是肿瘤进化的源泉这一观点。本综述讨论了量化表观基因组异质性和探索其各种原因和后果的最佳做法,包括表观基因组重编程、随机变化和持久记忆。此外,还讨论了限制表观基因组异质性的新治疗方法的设计问题,其长期目标是限制癌症的发展和恶化。本文章由计算机程序翻译,如有差异,请以英文原文为准。
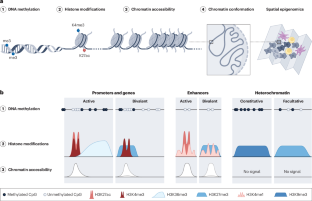

Epigenomic heterogeneity as a source of tumour evolution
In the past decade, remarkable progress in cancer medicine has been achieved by the development of treatments that target DNA sequence variants. However, a purely genetic approach to treatment selection is hampered by the fact that diverse cell states can emerge from the same genotype. In multicellular organisms, cell-state heterogeneity is driven by epigenetic processes that regulate DNA-based functions such as transcription; disruption of these processes is a hallmark of cancer that enables the emergence of defective cell states. Advances in single-cell technologies have unlocked our ability to quantify the epigenomic heterogeneity of tumours and understand its mechanisms, thereby transforming our appreciation of how epigenomic changes drive cancer evolution. This Review explores the idea that epigenomic heterogeneity and plasticity act as a reservoir of cell states and therefore as a source of tumour evolution. Best practices to quantify epigenomic heterogeneity and explore its various causes and consequences are discussed, including epigenomic reprogramming, stochastic changes and lasting memory. The design of new therapeutic approaches to restrict epigenomic heterogeneity, with the long-term objective of limiting cancer development and progression, is also addressed. Single-cell epigenomic technologies are refining our understanding of cancer evolution. Here Laisné et al. describe how epigenomic heterogeneity generates dynamic reservoirs of tumour cell states, through epigenomic reprogramming and selection among stochastic changes, which can be leveraged in the design of novel therapies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Reviews Cancer
医学-肿瘤学
CiteScore
111.90
自引率
0.40%
发文量
97
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Cancer, a part of the Nature Reviews portfolio of journals, aims to be the premier source of reviews and commentaries for the scientific communities it serves. The correct abbreviation for abstracting and indexing purposes is Nat. Rev. Cancer. The international standard serial numbers (ISSN) for Nature Reviews Cancer are 1474-175X (print) and 1474-1768 (online). Unlike other journals, Nature Reviews Cancer does not have an external editorial board. Instead, all editorial decisions are made by a team of full-time professional editors who are PhD-level scientists. The journal publishes Research Highlights, Comments, Reviews, and Perspectives relevant to cancer researchers, ensuring that the articles reach the widest possible audience due to their broad scope.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


