CRABP2 与细胞周期蛋白 D3 特异性结合的结构要求
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
细胞视黄酸结合蛋白 2(CRABP2)将视黄酸从细胞质转运至细胞核,然后将其货物转运至含有视黄酸受体的复合物,从而激活基因转录。我们利用纯化的蛋白质证明,CRABP2 也是一种细胞周期蛋白 D3 特异性结合蛋白,而且 CRABP2 的细胞周期蛋白 D3 结合位点与提出的 CRABP2 核定位序列重叠。这两个序列都位于螺旋-环-螺旋基团内,该基团形成了维甲酸结合口袋的盖子。该序列中的突变会阻止细胞周期蛋白 D3 和视黄酸的结合,从而促进 CRABP2 结构的形成,在这种结构中,视黄酸结合袋被另一种盖构象所占据。对 CRABP2 和细胞周期蛋白 D3 突变体的结构和功能分析,结合 CDK4/6- 细胞周期蛋白 D3-CRABP2 三元复合物的 AlphaFold 模型,支持在细胞周期蛋白 D3 C 端细胞周期蛋白盒折叠上确定一个 α 螺旋蛋白结合位点。本文章由计算机程序翻译,如有差异,请以英文原文为准。
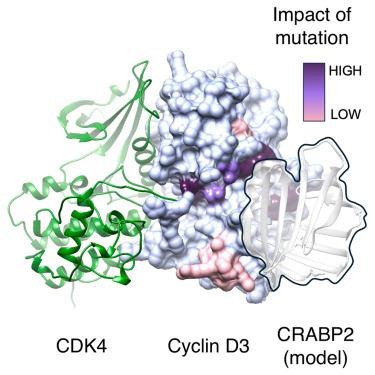
Structural requirements for the specific binding of CRABP2 to cyclin D3
Cellular retinoic acid binding protein 2 (CRABP2) transports retinoic acid from the cytoplasm to the nucleus where it then transfers its cargo to retinoic acid receptor-containing complexes leading to activation of gene transcription. We demonstrate using purified proteins that CRABP2 is also a cyclin D3-specific binding protein and that the CRABP2 cyclin D3 binding site and the proposed CRABP2 nuclear localization sequence overlap. Both sequences are within the helix-loop-helix motif that forms a lid to the retinoic acid binding pocket. Mutations within this sequence that block both cyclin D3 and retinoic acid binding promote formation of a CRABP2 structure in which the retinoic acid binding pocket is occupied by an alternative lid conformation. Structural and functional analysis of CRABP2 and cyclin D3 mutants combined with AlphaFold models of the ternary CDK4/6-cyclin D3-CRABP2 complex supports the identification of an α-helical protein binding site on the cyclin D3 C-terminal cyclin box fold.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


