单细胞转录组分析揭示幽门螺杆菌相关胃肿瘤发生过程中的细胞复杂性和微环境
IF 11.4
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
导言幽门螺杆菌(H. pylori)感染是胃癌(GC)的主要风险因素。目的我们制作了胃肿瘤发生的单细胞图谱,包括18个胃炎、胃肠化生(IM)和有或无幽门螺杆菌感染的胃癌标本。方法进行了单细胞RNA测序(scRNA-seq)。免疫荧光、免疫组织化学和 qRT-PCR 分析应用于第二个人类胃组织队列进行验证。结果单细胞 RNA 图谱突显了胃癌进展过程中的细胞异质性和组织生态学改变。不同的细胞系表现出独特的癌症相关表达谱,如肿瘤样上皮细胞亚群(EPC)、炎性癌相关成纤维细胞(iCAFs)和肿瘤相关巨噬细胞(TAM)。值得注意的是,我们发现来自癌前病变 GIM 的特定上皮亚群肠道细胞表现出与脂质代谢相关基因的高表达,而 HNF4G 被认为是其特定的转录因子。此外,我们还发现了幽门螺杆菌阳性和阴性上皮细胞、成纤维细胞和髓样细胞中表达不同的基因。此外,幽门螺杆菌阳性标本表现出丰富的细胞间通讯,其特征是 TNF、SPP1 和 THY1 信号网络显著活跃。本文章由计算机程序翻译,如有差异,请以英文原文为准。
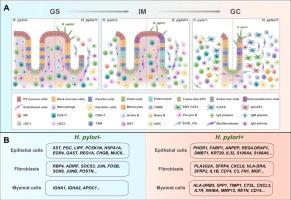
Single-cell transcriptomic profiling uncovers cellular complexity and microenvironment in gastric tumorigenesis associated with Helicobacter pylori
Introduction
Helicobacter pylori (H. pylori) infection is the main risk for gastric cancer (GC). However, the cellular heterogeneity and underlying molecular mechanisms in H. pylori-driven gastric tumorigenesis are poorly understood.Objective
Here, we generated a single-cell atlas of gastric tumorigenesis comprising 18 specimens of gastritis, gastric intestinal metaplasia (IM) and GC with or without H. pylori infection.Methods
Single-cell RNA sequencing (scRNA-seq) was performed. Immunofluorescence, immunohistochemistry and qRT-PCR analysis were applied in a second human gastric tissues cohort for validation. Bioinformatics analyses of public TCGA and GEO datasets were applied.Results
Single-cell RNA profile highlights cellular heterogeneity and alterations in tissue ecology throughout the progression of gastric carcinoma. Various cell lineages exhibited unique cancer-associated expression profiles, such as tumor-like epithelial cell subset (EPC), inflammatory cancer-associated fibroblasts (iCAFs) and Tumor-associated macrophage (TAM). Notably, we revealed that the specific epithelial subset enterocytes from the precancerous lesion GIM, exhibited elevated expression of genes related to lipid metabolism, and HNF4G was predicted as its specific transcription factor. Furthermore, we identified differentially expressed genes in H. pylori-positive and negative epithelial cells, fibroblasts and myeloid cells were identified. Futhermore, H. pylori-positive specimens exhibited enriched cell–cell communication, characterized by significantly active TNF, SPP1, and THY1 signaling networks.Conclusions
Our study provides a comprehensive landscape of the gastric carcinogenesis ecosystem and novel insights into the molecular mechanisms of different cell types in H. pylori-induced GC.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


