PARG 抑制可诱导 PARylated PARP1 的核聚集
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
聚(ADP-核糖)糖水解酶(PARG)抑制剂目前正处于临床开发阶段,用于治疗 DNA 修复缺陷型癌症;然而,其确切的作用机制仍不清楚。在这里,我们报告了 PARG 抑制会导致过多的 PARylated 多(ADP-核糖)聚合酶 1(PARP1),从而降低 PARP1 在 DNA 损伤位点正确定位的能力。令人震惊的是,定位错误的 PARP1 会在整个细胞核内聚集。削弱 PARP1 的催化活性可防止聚集体的形成,这表明 PAR 链在这一过程中起着关键作用。最后,我们发现 PARP1 核聚集体具有高度持久性,并与细胞质中被裂解的 PARP1 相关联,最终导致细胞死亡。总之,我们的数据揭示了 PARG 抑制剂细胞毒性的一种意想不到的机制,这将为这些药物作为抗癌疗法提供启示。本文章由计算机程序翻译,如有差异,请以英文原文为准。
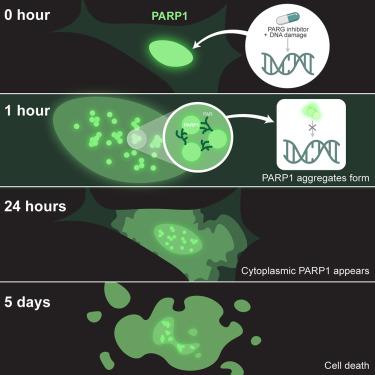
PARG inhibition induces nuclear aggregation of PARylated PARP1
Poly (ADP-ribose) glycohydrolase (PARG) inhibitors are currently under clinical development for the treatment of DNA repair-deficient cancers; however, their precise mechanism of action is still unclear. Here, we report that PARG inhibition leads to excessive PARylated poly (ADP-ribose) polymerase 1 (PARP1) reducing the ability of PARP1 to properly localize to sites of DNA damage. Strikingly, the mis-localized PARP1 accumulates as aggregates throughout the nucleus. Abrogation of the catalytic activity of PARP1 prevents aggregate formation, indicating that PAR chains play a key role in this process. Finally, we find that PARP1 nuclear aggregates were highly persistent and were associated with cleaved cytoplasmic PARP1, ultimately leading to cell death. Overall, our data uncover an unexpected mechanism of PARG inhibitor cytotoxicity, which will shed light on the use of these drugs as anti-cancer therapeutics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


