ML 增强的过氧化物酶体能力实现了多酶途径的区隔化
IF 13.7
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
重新利用细胞器进行专门代谢,为酿酒酵母等可发酵的单细胞生物提供了模仿不同植物组织内代谢途径分区的途径。过氧物酶体是极具吸引力的细胞器,因为酵母在葡萄糖上生长时不需要过氧物酶体,而且过氧物酶体可以有效地分隔异源酶,从而实现细胞膜原生代谢和过氧物酶体工程代谢的物理分离。然而,当不需要时,过氧物酶体受到抑制,导致异源蛋白的功能能力低下。在这里,我们设计了具有更强功能能力的过氧化物酶体,目的是将多达八种代谢酶分隔开来,以提高滴度。我们采用了一种机器学习方法,可以识别要过量表达的因子,最终使过氧物酶体的功能能力比野生型菌株提高了 137%。改进的途径区隔使单萜类香叶醇的生物合成滴度提高了 80%,达到 9.5 g L-1。本文章由计算机程序翻译,如有差异,请以英文原文为准。

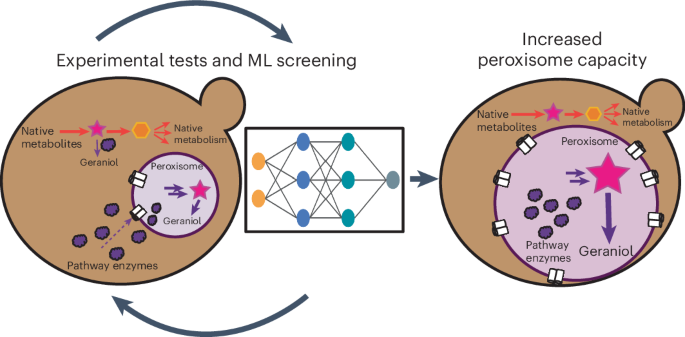
ML-enhanced peroxisome capacity enables compartmentalization of multienzyme pathway
Repurposing an organelle for specialized metabolism provides an avenue for fermentable, unicellular organisms such as Saccharomyces cerevisiae to mimic compartmentalization of metabolic pathways within different plant tissues. Peroxisomes are attractive organelles for repurposing as they are not required for yeast viability when grown on glucose and can efficiently compartmentalize heterologous enzymes to enable physical separation of cytosolic native metabolism and peroxisomal engineered metabolism. However, when not required, peroxisomes are repressed, leading to low functional capacities for heterologous proteins. Here we engineer peroxisomes with enhanced functional capacities, with the goal of compartmentalizing up to eight metabolic enzymes to enhance titers. We implement a machine learning pipeline that allows the identification of factors to overexpress, culminating in a 137% increase in peroxisome functional capacity compared to a wild-type strain. Improved pathway compartmentalization enables an 80% increase in the biosynthesis titers of the monoterpene geraniol, up to 9.5 g L−1. Metabolic engineering often suffers from competition between heterologous enzymes and native metabolism. Here the authors improve on a strategy of compartmentalizing metabolic pathways to the peroxisome by increasing peroxisome functional capacity, aided by machine learning, to produce geraniol.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature chemical biology
生物-生化与分子生物学
CiteScore
23.90
自引率
1.40%
发文量
238
审稿时长
12 months
期刊介绍:
Nature Chemical Biology stands as an esteemed international monthly journal, offering a prominent platform for the chemical biology community to showcase top-tier original research and commentary. Operating at the crossroads of chemistry, biology, and related disciplines, chemical biology utilizes scientific ideas and approaches to comprehend and manipulate biological systems with molecular precision.
The journal embraces contributions from the growing community of chemical biologists, encompassing insights from chemists applying principles and tools to biological inquiries and biologists striving to comprehend and control molecular-level biological processes. We prioritize studies unveiling significant conceptual or practical advancements in areas where chemistry and biology intersect, emphasizing basic research, especially those reporting novel chemical or biological tools and offering profound molecular-level insights into underlying biological mechanisms.
Nature Chemical Biology also welcomes manuscripts describing applied molecular studies at the chemistry-biology interface due to the broad utility of chemical biology approaches in manipulating or engineering biological systems. Irrespective of scientific focus, we actively seek submissions that creatively blend chemistry and biology, particularly those providing substantial conceptual or methodological breakthroughs with the potential to open innovative research avenues. The journal maintains a robust and impartial review process, emphasizing thorough chemical and biological characterization.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


