绘制癌症进展过程中染色体外 DNA 扩增的图谱
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
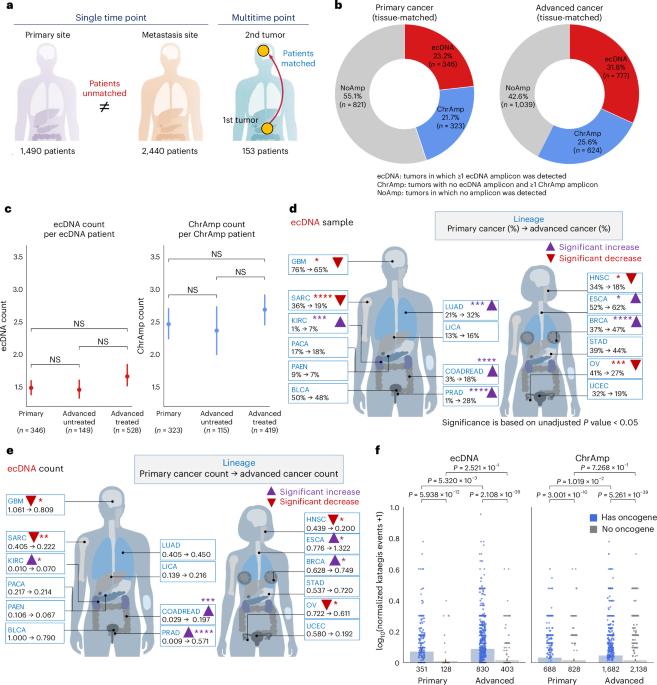
为了了解染色体外DNA(ecDNA)扩增在癌症进展中的作用,我们检测了8060例新诊断的原发性癌症、未治疗的转移瘤和重度预处理肿瘤中的病灶扩增并对其进行了分类。与新诊断的癌症相比,在未经治疗的转移瘤和预处理肿瘤中检测到的 ecDNAs 频率明显更高。与未经治疗的癌症相比,化疗预处理患者的肿瘤显示出更高的 ecDNA 频率。特别是,微管蛋白抑制与 ecDNA 的增加有关,表明 ecDNA 在治疗反应中的作用。在纵向匹配的肿瘤样本中,与染色体扩增相比,ecDNA更有可能被保留下来。ecDNA在不同时间点之间共享,晚期癌症中的ecDNA与私人ecDNA和新诊断肿瘤中的ecDNA相比,更有可能携带局部高突变事件。ecDNA局部高突变的变异等位基因比例相对较高,这与早期ecDNA突变有关。我们的研究结果表明,ecDNA 在癌症进展和转移过程中为肿瘤提供了竞争优势。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Mapping extrachromosomal DNA amplifications during cancer progression
To understand the role of extrachromosomal DNA (ecDNA) amplifications in cancer progression, we detected and classified focal amplifications in 8,060 newly diagnosed primary cancers, untreated metastases and heavily pretreated tumors. The ecDNAs were detected at significantly higher frequency in untreated metastatic and pretreated tumors compared to newly diagnosed cancers. Tumors from chemotherapy-pretreated patients showed significantly higher ecDNA frequency compared to untreated cancers. In particular, tubulin inhibition associated with ecDNA increases, suggesting a role for ecDNA in treatment response. In longitudinally matched tumor samples, ecDNAs were more likely to be retained compared to chromosomal amplifications. EcDNAs shared between time points, and ecDNAs in advanced cancers were more likely to harbor localized hypermutation events compared to private ecDNAs and ecDNAs in newly diagnosed tumors. Relatively high variant allele fractions of ecDNA localized hypermutations implicated early ecDNA mutagenesis. Our findings nominate ecDNAs to provide tumors with competitive advantages during cancer progression and metastasis. A pan-cancer genomic analysis finds an increase of extrachromosomal DNA (ecDNA) in treated and metastatic tumors compared to primary, untreated samples, as well as ecDNA features enriched in advanced disease.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


