通过转录组和 LC-MS/MS 分析鉴定稻纵卷叶螟的唾液蛋白。
IF 3.2
2区 农林科学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
咀嚼昆虫口腔分泌物(OS)中的唾液蛋白在昆虫取食期间与植物的相互作用中起着至关重要的作用。稻纵卷叶螟(Cnaphalocrocis medinalis)是全球水稻生产中臭名昭著的害虫,在取食过程中会引发防御反应,但人们对其唾液蛋白知之甚少。在这项研究中,我们证实褐飞虱在取食过程中会释放唾液蛋白。通过采用转录组分析和液相色谱-串联质谱(LC-MS/MS)技术,我们检测了来自中脊蝇唇唾液腺的唾液蛋白和来自其肠道的OS。共有 14,397 个基因在 RNA 水平上表达,并鉴定出 229 种唾液蛋白。与其他 25 个节肢动物物种的比较分析表明,有 43 种蛋白质是 C. medinalis 特有的。表达模式分析显示,大多数所选基因在肠道和幼虫阶段(4-5龄)高度表达。这些发现为未来唾液蛋白的功能研究提供了一个全面的资源,为了解 C. medinalis 调节植物防御能力的分子机制以及在害虫管理中的潜在应用提供了新的视角。本文章由计算机程序翻译,如有差异,请以英文原文为准。
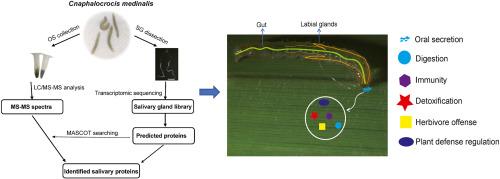
Identification of salivary proteins in the rice leaf folder Cnaphalocrocis medinalis by transcriptome and LC-MS/MS analyses
Salivary proteins in the oral secretion (OS) of chewing insects play a crucial role in insect-plant interactions during feeding. The rice leaf folder Cnaphalocrocis medinalis, a notorious pest in global rice production, triggers defense responses during feeding, but little is known about its salivary proteins. In this study, we confirmed that C. medinalis releases OS during feeding. By employing transcriptomic analysis and liquid chromatography-tandem mass spectroscopy (LC-MS/MS), we examined the salivary proteins from labial salivary glands and OS from C. medinalis. A total of 14,397 genes were expressed at the RNA level and 229 salivary proteins were identified. Comparative analysis with other 25 arthropod species revealed that 43 proteins were unique to C. medinalis. Expression pattern analysis revealed that most of the selected genes were highly expressed in the gut and the larval stages (4th–5th instar). These findings provide a comprehensive resource for future functional studies of salivary proteins, offering new insights into the molecular mechanisms by which C. medinalis modulates plant defenses and potential applications in pest management.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.40
自引率
5.30%
发文量
105
审稿时长
40 days
期刊介绍:
This international journal publishes original contributions and mini-reviews in the fields of insect biochemistry and insect molecular biology. Main areas of interest are neurochemistry, hormone and pheromone biochemistry, enzymes and metabolism, hormone action and gene regulation, gene characterization and structure, pharmacology, immunology and cell and tissue culture. Papers on the biochemistry and molecular biology of other groups of arthropods are published if of general interest to the readership. Technique papers will be considered for publication if they significantly advance the field of insect biochemistry and molecular biology in the opinion of the Editors and Editorial Board.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


