化脓性链球菌中的羟基氨基甲酸酯苷元结合蛋白 FtsB 的配体杂合性的结构基础
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
致病细菌必须确保吸收铁等营养金属才能生长,这使得它们的进口系统成为开发新抗菌模式的诱人目标。在致病性化脓性链球菌中,铁吸收系统 FtsABCD 可转运由羟酰胺类嗜苷酸盐包裹的铁。然而,由于化脓性链球菌无法产生这些代谢物,因此这一途径的生物学和临床意义尚未得到解决。在这里,我们证明了质膜周围结合蛋白 FtsB 不仅能识别羟氨酸盐类苷酸亚铁铬,还能识别铁氧胺 E(FOE)、铁氧胺 B(FOB)和双ucaberin(BIS),而且每种苷酸的亲和力都很高(nM 级)。通过 CH-π 相互作用,结合袋中多达七个芳香残基可容纳不同苷元的不同骨架,从而解释了配体的混杂性。总之,我们的观察结果揭示了化脓性链球菌如何利用其他微生物产生的各种嗜硒酸盐作为铁源,以确保这种珍贵的营养物质。本文章由计算机程序翻译,如有差异,请以英文原文为准。
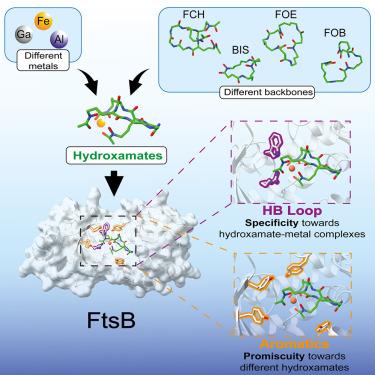
Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes
Pathogenic bacteria must secure the uptake of nutritional metals such as iron for their growth, making their import systems attractive targets for the development of new antimicrobial modalities. In the pathogenic bacterium Streptococcus pyogenes, the iron uptake system FtsABCD transports iron encapsulated by siderophores of the hydroxamate class. However, the inability of S. pyogenes to produce these metabolites makes the biological and clinical relevance of this route unresolved. Herein, we demonstrated that the periplasmic binding protein FtsB recognizes not only the hydroxamate siderophore ferrichrome, as previously documented, but also ferrioxamine E (FOE), ferrioxamine B (FOB), and bisucaberin (BIS), each of them with high affinity (nM level). Up to seven aromatic residues in the binding pocket accommodate the variable backbones of the different siderophores through CH-π interactions, explaining ligand promiscuity. Collectively, our observations revealed how S. pyogenes exploits the diverse xenosiderophores produced by other microorganisms as iron sources to secure this precious nutrient.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


