扩大分类群特异性微生物特征,预测农业系统中群落水平的微生物活动
IF 9.8
1区 农林科学
Q1 SOIL SCIENCE
引用次数: 0
摘要
土壤微生物发挥着许多重要的生态系统功能,包括氮(N)循环,它决定着农业生态系统中植物的生产力。尽管这些群落非常重要,但将微生物组成与生态系统功能联系起来一直是一个长期的挑战。利用定量稳定同位素探针(qSIP)测量的分类群特异性基质同化特征,可能为从微生物群落组成到群落级过程速率的扩展提供一种方法。为了测试扩大分类群特异性氮同化作用以预测群落级碳矿化率、氮矿化率和氮固定化率的可能性,我们在六个不同农场的土壤中使用 15N qSIP 测量了土壤特性、微生物活动和氮同化作用。以 DNA 15N 富集度衡量的氮同化作用在不同农场的分类群之间和分类群内部都存在差异。分类群的特定氮同化量经过汇总,计算出群落加权平均值,再与微生物生物量的测量值相结合,估算出新的微生物生物量氮产量。新微生物生物量的估计值反映了培养期间活跃微生物的生长情况,并与微生物活动有关。新产生的微生物生物量氮可预测土壤碳和氮的矿化率,可解释六种耕作制度中观察到的变化的 37-47%。这种方法强调了基于性状的方法将微生物群落结构数据与微生物介导的功能过程速率联系起来的能力。这种进步可能会提高我们理解和管理自然和农业生态系统中微生物介导过程(如氮循环)的能力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
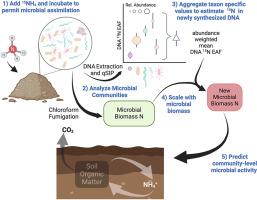
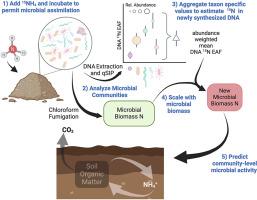
Scaling up taxon-specific microbial traits to predict community-level microbial activity in agricultural systems
Soil microorganisms perform many important ecosystem functions including nitrogen (N) cycling which dictates plant productivity in agricultural ecosystems. Despite the importance of these communities, connecting microbial composition with ecosystem function has been a long standing challenge. Taxon-specific substrate assimilation traits, measured with quantitative stable isotope probing (qSIP), may provide a means to scale from microbial community composition to community-level process rates. To test the potential for scaling up taxon-specific N assimilation to predict community-level rates of carbon mineralization, N mineralization, and N immobilization we measured soil properties, microbial activity, and N assimilation using 15N qSIP in soils from six distinct farm systems. N assimilation, measured as DNA 15N enrichment, varied among taxa and within taxa across farms. Taxon specific N assimilation was aggregated to calculate a community-weighted mean, which when combined with measures of microbial biomass was used to estimate new microbial biomass N production. This estimate of new microbial biomass production reflects the growth of active microbes over the incubation period and related to microbial activity. The new microbial biomass N produced was predictive of soil C and N mineralization rates, explaining 37–47% of the observed variation across the six farming systems. This approach highlights the ability of trait-based methods to relate microbial community structure data to microbially mediated functional process rates. Such advances may enhance our ability to understand and manage microbially mediated processes, such as N cycling, in both natural and agricultural ecosystems.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Soil Biology & Biochemistry
农林科学-土壤科学
CiteScore
16.90
自引率
9.30%
发文量
312
审稿时长
49 days
期刊介绍:
Soil Biology & Biochemistry publishes original research articles of international significance focusing on biological processes in soil and their applications to soil and environmental quality. Major topics include the ecology and biochemical processes of soil organisms, their effects on the environment, and interactions with plants. The journal also welcomes state-of-the-art reviews and discussions on contemporary research in soil biology and biochemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


