PROTAC 介导的核受体激活而非降解揭示了复杂的配体-受体相互作用网络
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
蛋白质分解靶向嵌合体(PROTACs)是一种含有相关蛋白质配体和 E3 泛素配体的异功能分子,通过 E3 招募到目标蛋白质上诱导蛋白质降解。PROTAC连接体的微小变化都会产生严重后果,包括降解活性的丧失,但这种变化的结构机制尚不清楚。为了研究这一现象,我们筛选了不同靶向模式的 PROTAC,发现 dTAG-13 是异生物感应孕烷 X 受体(PXR)的激活剂,PXR 可杂乱地结合各种配体。对 dTAG-13 类似物和前体的表征揭示了 PXR 结合分子、连接体和 E3 配体之间的相互作用,这些相互作用改变了 PXR 的活性而不会导致降解。与前体配体结合的 PXR 配体结合域的晶体结构显示,配体引起了结合口袋的扭曲,并在与 E3 复合物形成不相容的区域出现了连接体刺穿蛋白质外部的隧道,这突出表明了连接体环境对 PROTAC 活性的影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
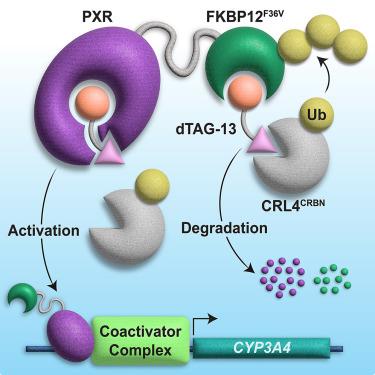
PROTAC-mediated activation, rather than degradation, of a nuclear receptor reveals complex ligand-receptor interaction network
Proteolysis-targeting chimeras (PROTACs) are heterobifunctional molecules containing a ligand for a protein of interest linked to an E3 ubiquitin ligase ligand that induce protein degradation through E3 recruitment to the target protein. Small changes in PROTAC linkers can have drastic consequences, including loss of degradation activity, but the structural mechanisms governing such changes are unclear. To study this phenomenon, we screened PROTACs of diverse targeting modalities and identified dTAG-13 as an activator of the xenobiotic-sensing pregnane X receptor (PXR), which promiscuously binds various ligands. Characterization of dTAG-13 analogs and precursors revealed interplay between the PXR-binding moiety, linker, and E3 ligand that altered PXR activity without inducing degradation. A crystal structure of PXR ligand binding domain bound to a precursor ligand showed ligand-induced binding pocket distortions and a linker-punctured tunnel to the protein exterior at a region incompatible with E3 complex formation, highlighting the effects of linker environment on PROTAC activity.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


