引导癌症中 mRNA 剪接的研究向临床转化
IF 66.8
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
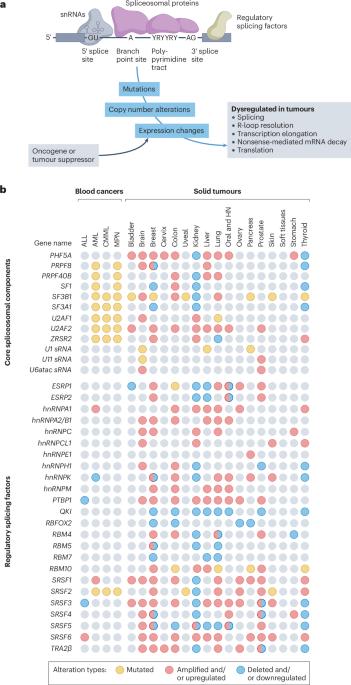
在几种类型的血液和实体恶性肿瘤中,剪接因子受到反复发生的体细胞突变和拷贝数变异的影响,这通常被视为剪接畸变可驱动癌症发生和发展的初步证据。然而,许多剪接体成分也 "兼职 "参与 DNA 修复和其他细胞过程,因此它们在癌症中的确切作用难以确定。不过,几乎没有人会否认,mRNA剪接失调是大多数癌症的普遍特征。正确解读这些分子指纹可以揭示肿瘤的新弱点和尚未开发的治疗机会。然而,多种技术挑战、挥之不去的误解和悬而未决的问题阻碍了临床转化。首先,由于短读RNA测序不擅长解析完整的mRNA异构体,以及长读RNA测序固有的浅读深度,尤其是在单细胞水平,因此癌症中剪接畸变的总体情况还没有得到很好的界定。虽然已知个别癌症相关同工酶会导致癌症进展,但广泛的剪接改变可能是人类癌症的一个同样重要的特征,而且可能更容易采取行动。这就是说,除了 "修复 "错误剪接的转录本之外,可能的治疗途径还包括利用小分子剪接体抑制剂加剧剪接畸变,利用合成致死方法靶向复发性剪接畸变,以及训练免疫系统识别剪接衍生的新抗原。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Steering research on mRNA splicing in cancer towards clinical translation
Splicing factors are affected by recurrent somatic mutations and copy number variations in several types of haematologic and solid malignancies, which is often seen as prima facie evidence that splicing aberrations can drive cancer initiation and progression. However, numerous spliceosome components also ‘moonlight’ in DNA repair and other cellular processes, making their precise role in cancer difficult to pinpoint. Still, few would deny that dysregulated mRNA splicing is a pervasive feature of most cancers. Correctly interpreting these molecular fingerprints can reveal novel tumour vulnerabilities and untapped therapeutic opportunities. Yet multiple technological challenges, lingering misconceptions, and outstanding questions hinder clinical translation. To start with, the general landscape of splicing aberrations in cancer is not well defined, due to limitations of short-read RNA sequencing not adept at resolving complete mRNA isoforms, as well as the shallow read depth inherent in long-read RNA-sequencing, especially at single-cell level. Although individual cancer-associated isoforms are known to contribute to cancer progression, widespread splicing alterations could be an equally important and, perhaps, more readily actionable feature of human cancers. This is to say that in addition to ‘repairing’ mis-spliced transcripts, possible therapeutic avenues include exacerbating splicing aberration with small-molecule spliceosome inhibitors, targeting recurrent splicing aberrations with synthetic lethal approaches, and training the immune system to recognize splicing-derived neoantigens. Although splicing factors are altered in cancer through mutations and copy number variations, their exact role remains challenging to define. In this Roadmap, Anczukow, Thomas-Tikhonenko and colleagues explore recent advances in splicing biology and provide guidance on leveraging these insights to facilitate the clinical application of compounds that target or exploit aberrant splicing patterns in cancer.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Reviews Cancer
医学-肿瘤学
CiteScore
111.90
自引率
0.40%
发文量
97
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Cancer, a part of the Nature Reviews portfolio of journals, aims to be the premier source of reviews and commentaries for the scientific communities it serves. The correct abbreviation for abstracting and indexing purposes is Nat. Rev. Cancer. The international standard serial numbers (ISSN) for Nature Reviews Cancer are 1474-175X (print) and 1474-1768 (online). Unlike other journals, Nature Reviews Cancer does not have an external editorial board. Instead, all editorial decisions are made by a team of full-time professional editors who are PhD-level scientists. The journal publishes Research Highlights, Comments, Reviews, and Perspectives relevant to cancer researchers, ensuring that the articles reach the widest possible audience due to their broad scope.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


