GeneCompass:利用知识型跨物种基础模型破译通用基因调控机制
IF 28.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
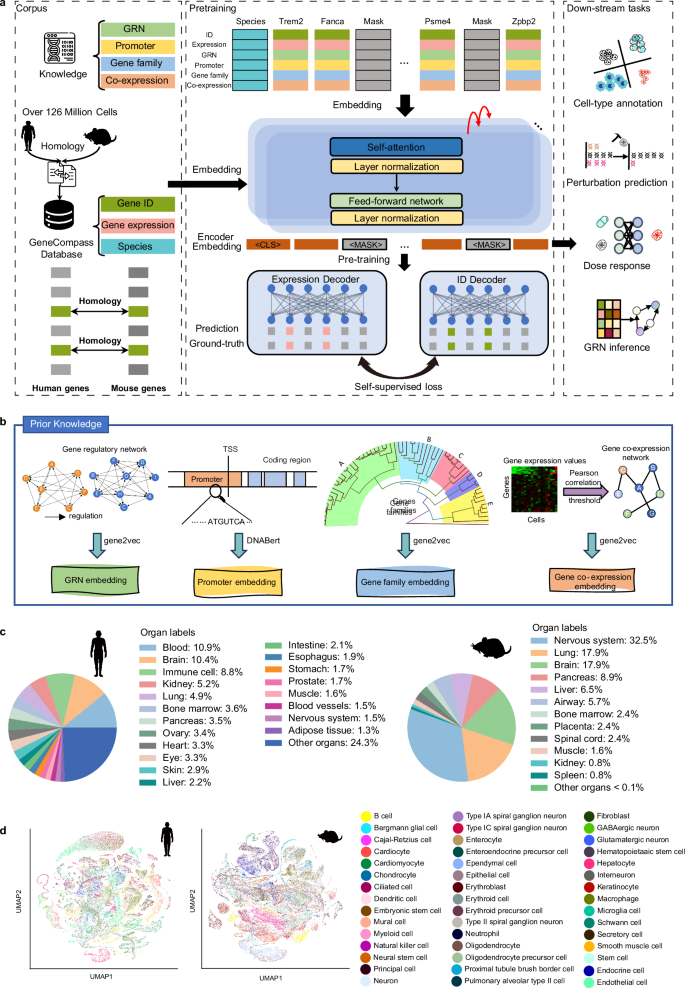
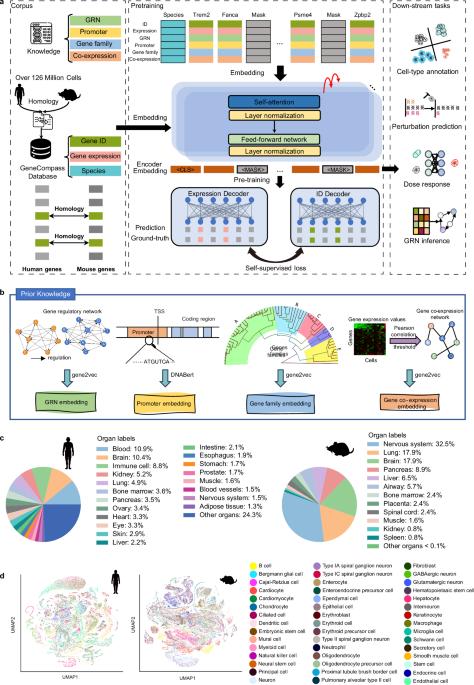
破译不同生物体中的通用基因调控机制,对于增进我们对基本生命过程的了解和促进临床应用具有巨大潜力。然而,传统的研究范式主要关注单个模式生物,并没有整合不同物种的各种细胞类型。单细胞测序和深度学习技术的最新突破为应对这一挑战提供了前所未有的机遇。在这项研究中,我们建立了一个包含超过 1.2 亿个人类和小鼠单细胞转录组的广泛数据集。经过数据预处理后,我们获得了 101,768,420 个单细胞转录组,并开发了一个基于知识的跨物种基础模型,命名为 GeneCompass。在预训练过程中,GeneCompass 有效地整合了四种先验生物学知识,以自我监督的方式增强了我们对基因调控机制的理解。通过对多个下游任务进行微调,GeneCompass 在单一物种的各种应用中表现优于最先进的模型,并开启了跨物种生物研究的新领域。我们还利用 GeneCompass 搜索与细胞命运转变相关的关键因素,结果表明预测的候选基因能成功诱导人类胚胎干细胞向性腺命运分化。总之,GeneCompass 展示了利用人工智能技术破译通用基因调控机制的优势,并显示了加速发现关键细胞命运调控因子和候选药物靶点的巨大潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
GeneCompass: deciphering universal gene regulatory mechanisms with a knowledge-informed cross-species foundation model
Deciphering universal gene regulatory mechanisms in diverse organisms holds great potential for advancing our knowledge of fundamental life processes and facilitating clinical applications. However, the traditional research paradigm primarily focuses on individual model organisms and does not integrate various cell types across species. Recent breakthroughs in single-cell sequencing and deep learning techniques present an unprecedented opportunity to address this challenge. In this study, we built an extensive dataset of over 120 million human and mouse single-cell transcriptomes. After data preprocessing, we obtained 101,768,420 single-cell transcriptomes and developed a knowledge-informed cross-species foundation model, named GeneCompass. During pre-training, GeneCompass effectively integrated four types of prior biological knowledge to enhance our understanding of gene regulatory mechanisms in a self-supervised manner. By fine-tuning for multiple downstream tasks, GeneCompass outperformed state-of-the-art models in diverse applications for a single species and unlocked new realms of cross-species biological investigations. We also employed GeneCompass to search for key factors associated with cell fate transition and showed that the predicted candidate genes could successfully induce the differentiation of human embryonic stem cells into the gonadal fate. Overall, GeneCompass demonstrates the advantages of using artificial intelligence technology to decipher universal gene regulatory mechanisms and shows tremendous potential for accelerating the discovery of critical cell fate regulators and candidate drug targets.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


