遗传结构调和了复杂性状的关联研究和相关研究
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
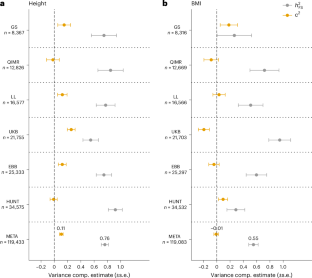
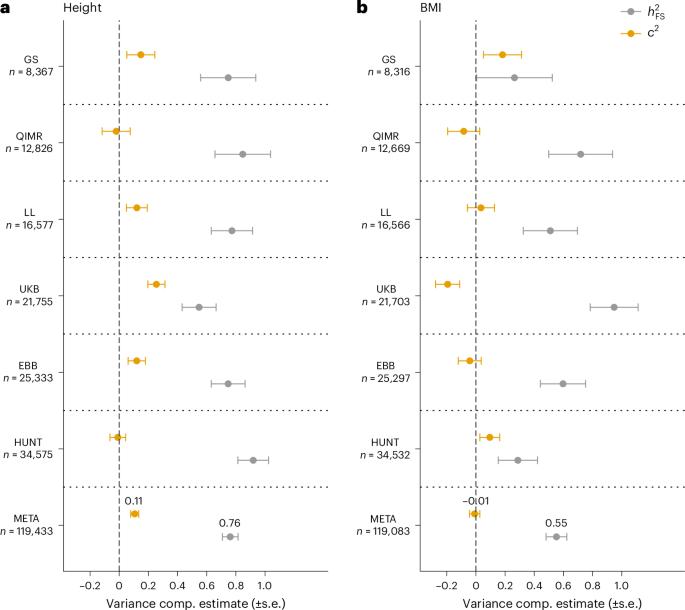
连锁研究成功地绘制了单基因遗传病的基因位点图,但在应用于常见疾病时却大多失败。相反,全基因组关联研究(GWAS)发现了数千个 SNP 与复杂性状之间可复制的关联,但只捕捉到不到一半的总遗传率。在本研究中,我们对这两种方法进行了调和,表明来自 119,000 对同胞的身高和体重指数(BMI)的关联信号与全基因组关联研究(GWAS)确定的位点具有共定位性。与多基因性一致,我们观察到以下几点:全基因组范围内的关联检验统计量膨胀;GWAS 结果可预测关联信号;根据多基因评分调整表型可减少关联信号。最后,我们开发了一种方法,利用同胞间的重组率分层、同源共享来无偏估计身高(0.76 ± 0.05)和体重指数(0.55 ± 0.07)的遗传率。我们的研究结果表明,GWAS 确定的基因位点仍有很大的遗传率没有考虑在内,而这种残余遗传变异是多基因的,并且富集在这些基因位点附近。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genetic architecture reconciles linkage and association studies of complex traits
Linkage studies have successfully mapped loci underlying monogenic disorders, but mostly failed when applied to common diseases. Conversely, genome-wide association studies (GWASs) have identified replicable associations between thousands of SNPs and complex traits, yet capture less than half of the total heritability. In the present study we reconcile these two approaches by showing that linkage signals of height and body mass index (BMI) from 119,000 sibling pairs colocalize with GWAS-identified loci. Concordant with polygenicity, we observed the following: a genome-wide inflation of linkage test statistics; that GWAS results predict linkage signals; and that adjusting phenotypes for polygenic scores reduces linkage signals. Finally, we developed a method using recombination rate-stratified, identity-by-descent sharing between siblings to unbiasedly estimate heritability of height (0.76 ± 0.05) and BMI (0.55 ± 0.07). Our results imply that substantial heritability remains unaccounted for by GWAS-identified loci and this residual genetic variation is polygenic and enriched near these loci. Analyses of height and body mass index in 119,000 sibling pairs show that linkage and genome-wide association signals colocalize. Further analyses suggest that family-based linkage signals are fully consistent with a highly polygenic architecture.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


