拉帕替尼作为流行性癌症辅助疗法的前瞻性作用:以表皮生长因子受体(EGFR)和表皮生长因子受体(HER2)为靶点的硅学分析透视。
IF 3
3区 生物学
Q3 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
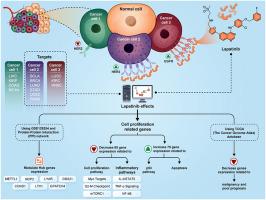
导言:各种证据表明,不同癌症中表皮生长因子受体(EGFR)和表皮生长因子受体(HER2)水平的升高会导致癌细胞的增殖、侵袭和转移。在本研究中,我们利用硅学数据对这两种受体在不同癌症中的表达变化进行了全面调查。此外,我们还研究了拉帕替尼作为这两种受体的抑制剂在各种癌症中的治疗潜力:方法:从癌症基因组图谱(TCGA)下载流行癌症的 RNAseq 数据。经过初步预处理后,对 HER2、表皮生长因子受体和候选基因(根据它们与表皮生长因子受体和 HER2 信号通路的关系确定)的表达变化进行了检测。人类蛋白质图谱数据用于评估 HER2 和表皮生长因子受体的蛋白质表达。利用 GSE129254 确定与拉帕替尼相关的分子通路和候选基因。蛋白-蛋白相互作用网络用于识别受拉帕替尼影响的枢纽基因。利用常见癌症的临床数据,通过 Cox 回归检验研究候选基因的表达与患者死亡率之间的相关性:结果:研究结果表明,在肾癌、肺癌、乳腺癌、膀胱癌、胰腺癌、头颈癌、胃癌和子宫内膜癌等癌症中,HER2和表皮生长因子受体(EGFR)的表达水平在mRNA和蛋白质水平上都有明显增加(p值<0.01)。此外,在某些癌症中,超过 30% 的样本显示 HER2 或表皮生长因子受体表达增加了两倍。对GSE129254数据的分析表明,拉帕替尼降低了许多与细胞增殖相关的基因的表达。METTL1、LYAR、LTV1、CCND1、NOP2和DDX21被确定为与拉帕替尼效应相关的枢纽基因。我们的研究结果表明,许多中枢基因在候选癌症中表达升高,其中一些基因的上调与预后不良相关:结论:我们的研究结果表明,在某些常见癌症中,HER2和表皮生长因子受体(EGFR)的表达水平上调,这表明拉帕替尼除用于治疗乳腺癌外,还可用于治疗这些癌症。此外,我们还证明,一些在常见癌症中表达增加并与预后不良有关的基因有可能受到拉帕替尼的调节。本文章由计算机程序翻译,如有差异,请以英文原文为准。
The Prospective role of lapatinib as an adjuvant therapy in prevalent cancers: Insights from in silico analysis targeting EGFR and HER2
Introduction
Various pieces of evidence suggest an elevation in the levels of EGFR and HER2 in different cancers leading to the proliferation, invasion, and metastasis of cancer cells. In this study, we conducted a comprehensive investigation into the expression alterations of these two receptors in various cancers using in silico data. In addition, we investigated the therapeutic potential of lapatinib as an inhibitor of these receptors in various cancer types.
Methods
RNAseq data for prevalent cancers were downloaded from The Cancer Genome Atlas (TCGA). After initial preprocessing, expression changes of HER2, EGFR, and candidate genes—identified based on their association with EGFR and HER2 signaling pathways—were examined. Human protein atlas data were utilized to assess the protein expression of HER2 and EGFR. GSE129254 was employed to identify molecular pathways and candidate genes associated with lapatinib. The protein-protein interaction network was used to identify lapatinib-influenced hub genes. Clinical data for common cancers were used to investigate the correlation between the expression of candidate genes and patients' mortality rates by Cox regression test.
Results
The findings clearly indicated a significant increase in the expression levels of HER2 and EGFR in cancers such as kidney, lung, breast, bladder, pancreas, head and neck, stomach, and endometrial, both at the mRNA and protein levels (p-value <0.01). Additionally, more than 30 % of samples in some cancers showed a twofold increase in HER2 or EGFR expression. The analysis of GSE129254 data revealed that lapatinib reduces the expression of numerous genes associated with cell proliferation. METTL1, LYAR, LTV1, CCND1, NOP2, and DDX21 were identified as hub genes related to the effect of lapatinib. Our results demonstrated that many hub genes exhibited elevated expression in candidate cancers, and the upregulation of some of them was correlated with poor prognosis.
Conclusion
Our results indicate an upregulation in the expression levels of HER2 and EGFR in certain common cancers, suggesting that lapatinib, in addition to breast cancer, could be considered for the treatment of these cancers. Furthermore, we demonstrated that some genes with increased expression in prevalent cancers and associated with poor prognosis have the potential to be modulated by lapatinib.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular and Cellular Probes
生物-生化研究方法
CiteScore
6.80
自引率
0.00%
发文量
52
审稿时长
16 days
期刊介绍:
MCP - Advancing biology through–omics and bioinformatic technologies wants to capture outcomes from the current revolution in molecular technologies and sciences. The journal has broadened its scope and embraces any high quality research papers, reviews and opinions in areas including, but not limited to, molecular biology, cell biology, biochemistry, immunology, physiology, epidemiology, ecology, virology, microbiology, parasitology, genetics, evolutionary biology, genomics (including metagenomics), bioinformatics, proteomics, metabolomics, glycomics, and lipidomics. Submissions with a technology-driven focus on understanding normal biological or disease processes as well as conceptual advances and paradigm shifts are particularly encouraged. The Editors welcome fundamental or applied research areas; pre-submission enquiries about advanced draft manuscripts are welcomed. Top quality research and manuscripts will be fast-tracked.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


