果蝇胚胎中成对规则蛋白质模式的建模和校准:从偶数跳和傅氏太郎到无翼表达网络。
IF 2.5
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
摘要
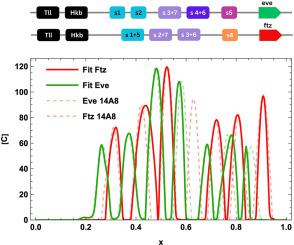
我们模拟并校准了在早期发育过程中建立的 Even-skipped(夏娃)和 Fushi-tarazu(Ftz)成对规则蛋白沿 Drosphila 胚胎前后轴的七条纹图案的分布。我们确定了五个夏娃增强子的推定抑制组合,并探索了夏娃和 Ftz 之间的互补模式关系。夏娃和 Ftz 的调控因子是条纹特异性 DNA 增强子,其激活率与胚胎位置有关,并受缺口蛋白家族的调控。我们实现了夏娃条纹模式的显著数据匹配,校准后的模型重现了间隙基因突变实验。我们提出了从 Eve 和 Ftz 增强子推断无翼虫(Wg)14 条纹模式的扩展工作,从而阐明了 Drosphila 早期发育过程中基因表达网络的层次结构。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Modelling and calibration of pair-rule protein patterns in Drosophila embryo: From Even-skipped and Fushi-tarazu to Wingless expression networks
We modelled and calibrated the distributions of the seven-stripe patterns of Even-skipped (Eve) and Fushi-tarazu (Ftz) pair-rule proteins along the anteroposterior axis of the Drosphila embryo, established during early development. We have identified the putative repressive combinations for five Eve enhancers, and we have explored the relationship between Eve and Ftz for complementary patterns. The regulators of Eve and Ftz are stripe-specific DNA enhancers with embryo position-dependent activation rates and are regulated by the gap family of proteins. We achieved remarkable data matching of the Eve stripe pattern, and the calibrated model reproduces gap gene mutation experiments. Extended work inferring the Wingless (Wg) fourteen stripe pattern from Eve and Ftz enhancers have been proposed, clarifying the hierarchical structure of Drosphila's genetic expression network during early development.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


