蛋白质表征:为生物催化中的机器学习编码生物信息。
IF 12.1
1区 工程技术
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
与传统化学相比,酶提供了一种更环保、影响更小的解决方案,但它们在工业环境中的应用往往需要额外的工程设计,这是一项具有挑战性且费力的工作。为了解决这个问题,可以利用机器学习的力量来生成预测模型,从而对改进的酶特性进行硅学研究和工程设计。然而,这种机器学习模型需要将复杂的生物信息转换为数字输入,也称为蛋白质表征。因此,在本综述中,我们将探讨将蛋白质信息编码为数字表征以用于机器学习的关键步骤。我们选择了最重要的方法来编码三种不同的生物蛋白质表征--主序列、三维结构和动力学--以探讨它们对就业和归纳偏差的要求。我们还介绍了蛋白质和底物的组合表征,将其作为生物催化中的新兴工具。我们建议将固定表征、基于规则的编码策略集合和从大型神经网络潜空间中提取的学习表征进行划分。为了选择最合适的蛋白质表征,我们提出了两个主要考虑因素。第一个因素是模型设置,它受到训练数据集大小和架构选择的影响。第二个因素是模型目标,如对检测属性的考虑、野生型模型与突变预测模型之间的差异以及对可解释性的要求。本综述旨在为在未来生物催化机器学习模型中正确表示酶提供信息和指导。本文章由计算机程序翻译,如有差异,请以英文原文为准。
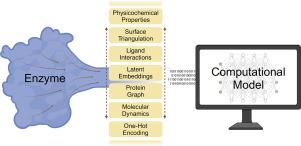
Protein representations: Encoding biological information for machine learning in biocatalysis
Enzymes offer a more environmentally friendly and low-impact solution to conventional chemistry, but they often require additional engineering for their application in industrial settings, an endeavour that is challenging and laborious. To address this issue, the power of machine learning can be harnessed to produce predictive models that enable the in silico study and engineering of improved enzymatic properties. Such machine learning models, however, require the conversion of the complex biological information to a numerical input, also called protein representations. These inputs demand special attention to ensure the training of accurate and precise models, and, in this review, we therefore examine the critical step of encoding protein information to numeric representations for use in machine learning. We selected the most important approaches for encoding the three distinct biological protein representations — primary sequence, 3D structure, and dynamics — to explore their requirements for employment and inductive biases. Combined representations of proteins and substrates are also introduced as emergent tools in biocatalysis. We propose the division of fixed representations, a collection of rule-based encoding strategies, and learned representations extracted from the latent spaces of large neural networks. To select the most suitable protein representation, we propose two main factors to consider. The first one is the model setup, which is influenced by the size of the training dataset and the choice of architecture. The second factor is the model objectives such as consideration about the assayed property, the difference between wild-type models and mutant predictors, and requirements for explainability. This review is aimed at serving as a source of information and guidance for properly representing enzymes in future machine learning models for biocatalysis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Biotechnology advances
工程技术-生物工程与应用微生物
CiteScore
25.50
自引率
2.50%
发文量
167
审稿时长
37 days
期刊介绍:
Biotechnology Advances is a comprehensive review journal that covers all aspects of the multidisciplinary field of biotechnology. The journal focuses on biotechnology principles and their applications in various industries, agriculture, medicine, environmental concerns, and regulatory issues. It publishes authoritative articles that highlight current developments and future trends in the field of biotechnology. The journal invites submissions of manuscripts that are relevant and appropriate. It targets a wide audience, including scientists, engineers, students, instructors, researchers, practitioners, managers, governments, and other stakeholders in the field. Additionally, special issues are published based on selected presentations from recent relevant conferences in collaboration with the organizations hosting those conferences.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


