快速设计噬菌体鸡尾酒,抑制肠道耐碳青霉烯类肺炎克雷伯氏菌的负担和毒力
IF 20.6
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
摘要
抗生素的使用会导致肠道微生物群中具有多重耐药性的病原菌增多,从而引发危及生命的感染。传统抗生素的选择性替代品亟待开发。在这里,我们描述了一个克雷伯氏菌噬菌体库(Klebsiella PhageBank),用于定制设计噬菌体鸡尾酒,以治疗多重耐药肺炎克雷伯氏菌。利用耐碳青霉烯类肺炎克雷伯氏菌的转座子文库,我们确定了主要克雷伯氏菌噬菌体家族中噬菌体感染所需的宿主因子。利用噬菌体库的多样性,我们配制出了噬菌体组合,能以最小的噬菌体耐药性消灭肺炎克雷伯氏菌。优化后的鸡尾酒选择性地抑制了小鼠肠道中肺炎克雷伯菌的负担,并促使作为噬菌体受体的关键毒力因子丧失。噬菌体介导的肠道细菌种群多样化导致了噬菌体变种的共同进化,这些变种具有更强的毒力和更广的宿主范围。总之,克雷伯氏菌噬菌体库为噬菌体疗法提供了一个路线图,以对付一种关键的耐多药人类病原体。本文章由计算机程序翻译,如有差异,请以英文原文为准。
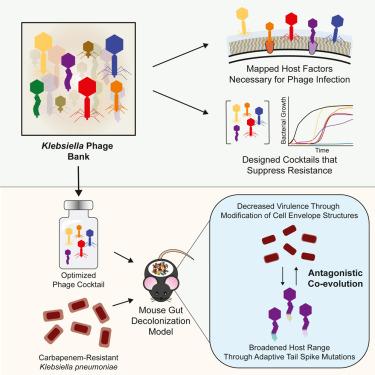
Rapid design of bacteriophage cocktails to suppress the burden and virulence of gut-resident carbapenem-resistant Klebsiella pneumoniae
Antibiotic use can lead to the expansion of multi-drug-resistant pathobionts within the gut microbiome that can cause life-threatening infections. Selective alternatives to conventional antibiotics are in dire need. Here, we describe a Klebsiella PhageBank for the tailored design of bacteriophage cocktails to treat multi-drug-resistant Klebsiella pneumoniae. Using a transposon library in carbapenem-resistant K. pneumoniae, we identify host factors required for phage infection in major Klebsiella phage families. Leveraging the diversity of the PhageBank, we formulate phage combinations that eliminate K. pneumoniae with minimal phage resistance. Optimized cocktails selectively suppress the burden of K. pneumoniae in the mouse gut and drive the loss of key virulence factors that act as phage receptors. Phage-mediated diversification of bacterial populations in the gut leads to co-evolution of phage variants with higher virulence and broader host range. Altogether, the Klebsiella PhageBank charts a roadmap for phage therapy against a critical multidrug-resistant human pathogen.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


