用于探索人脑发育过程中与神经系统疾病相关基因的细胞和时间特异性的综合单细胞图谱。
IF 9.5
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
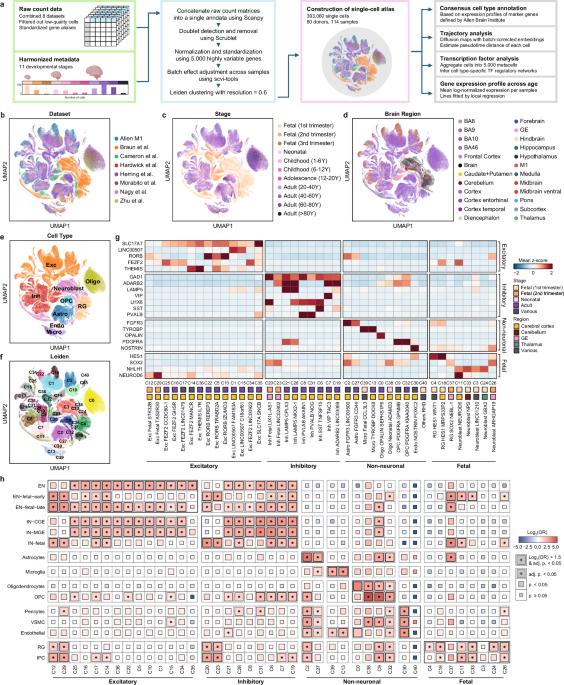
单细胞技术有助于对不同脑区进行广泛的转录组普查,从而增进了对人类大脑的全面了解。然而,由于发育上的差异,对神经系统疾病的细胞和时间特异性的理解仍然模糊不清。为了填补这一空白,我们通过整合多个单细胞 RNA 测序数据集来说明发育过程中疾病风险基因表达的动态变化。我们构建了一个全面的发育中人脑单细胞图谱,涵盖了不同发育阶段的393,060个单细胞。时间分析揭示了紊乱风险基因(包括与自闭症相关的基因)的独特表达模式,突出了它们在不同神经元和神经胶质细胞系中的时间调控。我们发现了在不同发育阶段出现分化的不同神经元系,每个神经元系都表现出失调相关基因的特定时间表达模式。通过分子图谱确定的非神经元细胞系也表现出时间特异性表达,这表明细胞成熟与失常风险之间存在联系。此外,我们还探索了大脑早期发育的调控机制,揭示了与神经元紊乱相关的胎儿细胞类型的丰富模式,表明产前阶段对疾病的决定性影响。我们的研究结果有助于对细胞类型与疾病的关联进行无偏见的比较,并深入了解风险基因在发育过程中的动态变化,为深入了解神经系统疾病铺平了道路。本文章由计算机程序翻译,如有差异,请以英文原文为准。
An integrative single-cell atlas for exploring the cellular and temporal specificity of genes related to neurological disorders during human brain development
Single-cell technologies have enhanced comprehensive knowledge regarding the human brain by facilitating an extensive transcriptomic census across diverse brain regions. Nevertheless, understanding the cellular and temporal specificity of neurological disorders remains ambiguous due to developmental variations. To address this gap, we illustrated the dynamics of disorder risk gene expression under development by integrating multiple single-cell RNA sequencing datasets. We constructed a comprehensive single-cell atlas of the developing human brain, encompassing 393,060 single cells across diverse developmental stages. Temporal analysis revealed the distinct expression patterns of disorder risk genes, including those associated with autism, highlighting their temporal regulation in different neuronal and glial lineages. We identified distinct neuronal lineages that diverged across developmental stages, each exhibiting temporal-specific expression patterns of disorder-related genes. Lineages of nonneuronal cells determined by molecular profiles also showed temporal-specific expression, indicating a link between cellular maturation and the risk of disorder. Furthermore, we explored the regulatory mechanisms involved in early brain development, revealing enriched patterns of fetal cell types associated with neuronal disorders indicative of the prenatal stage’s influence on disease determination. Our findings facilitate unbiased comparisons of cell type‒disorder associations and provide insight into dynamic alterations in risk genes during development, paving the way for a deeper understanding of neurological disorders. The growth of the human brain is a complicated process that begins before birth and continues into young adulthood. Researchers focused on how genes related to brain disorders are expressed in different cells over time. They gathered data from 114 human brain samples, creating a single-cell atlas that traces brain growth from early fetal stages to adulthood. The results showed distinct patterns of gene expression linked to disorders like autism and developmental delay, especially in neurons during early growth. The study also emphasized the role of glial cells in brain conditions, such as Alzheimer’s and Parkinson’s disease, by showing specific gene expression patterns in these cells related to the disorders. Researchers conclude that their single-cell atlas greatly improves our understanding of brain growth and the molecular mechanisms behind brain disorders. This summary was initially drafted using artificial intelligence, then revised and fact-checked by the author.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Experimental and Molecular Medicine
医学-生化与分子生物学
CiteScore
19.50
自引率
0.80%
发文量
166
审稿时长
3 months
期刊介绍:
Experimental & Molecular Medicine (EMM) stands as Korea's pioneering biochemistry journal, established in 1964 and rejuvenated in 1996 as an Open Access, fully peer-reviewed international journal. Dedicated to advancing translational research and showcasing recent breakthroughs in the biomedical realm, EMM invites submissions encompassing genetic, molecular, and cellular studies of human physiology and diseases. Emphasizing the correlation between experimental and translational research and enhanced clinical benefits, the journal actively encourages contributions employing specific molecular tools. Welcoming studies that bridge basic discoveries with clinical relevance, alongside articles demonstrating clear in vivo significance and novelty, Experimental & Molecular Medicine proudly serves as an open-access, online-only repository of cutting-edge medical research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


