种群特异性推定因果变异塑造数量性状
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
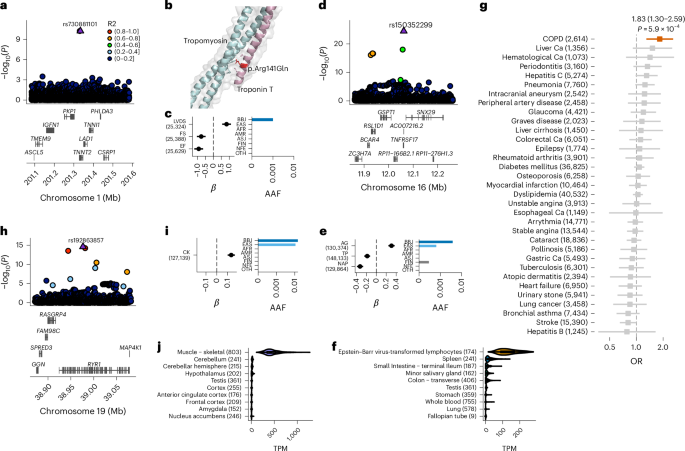
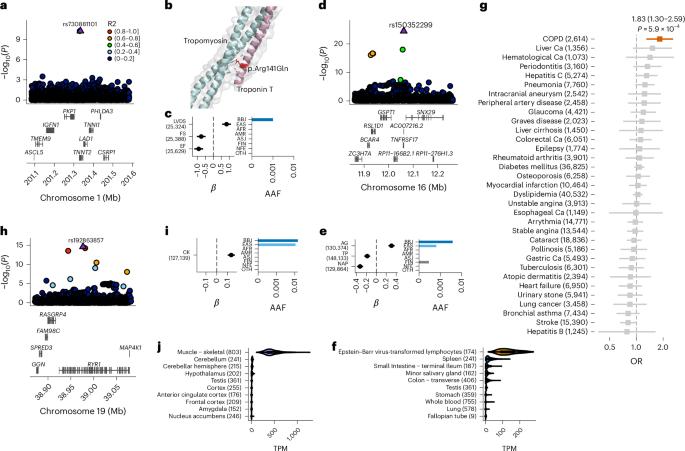
人类基因变异与许多性状相关,其机制在很大程度上是未知的。在这里,我们结合约 26 万名日本研究参与者、日本特异性基因型参考面板和统计精细图谱,在 63 个数量性状中发现了 4423 个重要位点,其中 601 个为新位点,9406 个为推测因果变异。新的关联包括日本特异性编码变异、剪接变异和非编码变异,例如 TNNT2 中的一个破坏性错义变异 rs730881101 与心脏功能降低和心力衰竭风险增加有关(P = 1.4 × 10-15 和几率比 = 4.5,95% 置信区间 = 3.1-6.5)。推定的非编码因果变异得到了最先进的硅学功能测试的支持,其效应大小与编码变异相当。因果变异新机制的一个可信例子是,3′非翻译区(UTR)中的因果变异富集,包括日本特异的 IL6 rs13306436,它与促炎症特征和结核病保护相关。我们的实验表明,带有rs13306436的转录本能抵抗RNA结合蛋白regnase-1对mRNA的降解。我们的研究提供了一份待测试功能的精细映射因果变异体列表,并强调了在不同人群中进行测序、基因分型和关联工作的重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Population-specific putative causal variants shape quantitative traits
Human genetic variants are associated with many traits through largely unknown mechanisms. Here, combining approximately 260,000 Japanese study participants, a Japanese-specific genotype reference panel and statistical fine-mapping, we identified 4,423 significant loci across 63 quantitative traits, among which 601 were new, and 9,406 putatively causal variants. New associations included Japanese-specific coding, splicing and noncoding variants, exemplified by a damaging missense variant rs730881101 in TNNT2 associated with lower heart function and increased risk for heart failure (P = 1.4 × 10−15 and odds ratio = 4.5, 95% confidence interval = 3.1–6.5). Putative causal noncoding variants were supported by state-of-art in silico functional assays and had comparable effect sizes to coding variants. A plausible example of new mechanisms of causal variants is an enrichment of causal variants in 3′ untranslated regions (UTRs), including the Japanese-specific rs13306436 in IL6 associated with pro-inflammatory traits and protection against tuberculosis. We experimentally showed that transcripts with rs13306436 are resistant to mRNA degradation by regnase-1, an RNA-binding protein. Our study provides a list of fine-mapped causal variants to be tested for functionality and underscores the importance of sequencing, genotyping and association efforts in diverse populations. Genome-wide association and fine-mapping analyses in approximately 260,000 Japanese individuals combined with a newly constructed Japanese-specific genotype reference panel identify hundreds of new loci and putative causal variants for 63 quantitative traits.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


