绝对定量和碱基分辨率测序揭示了假尿苷在人类转录组中的全面分布。
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
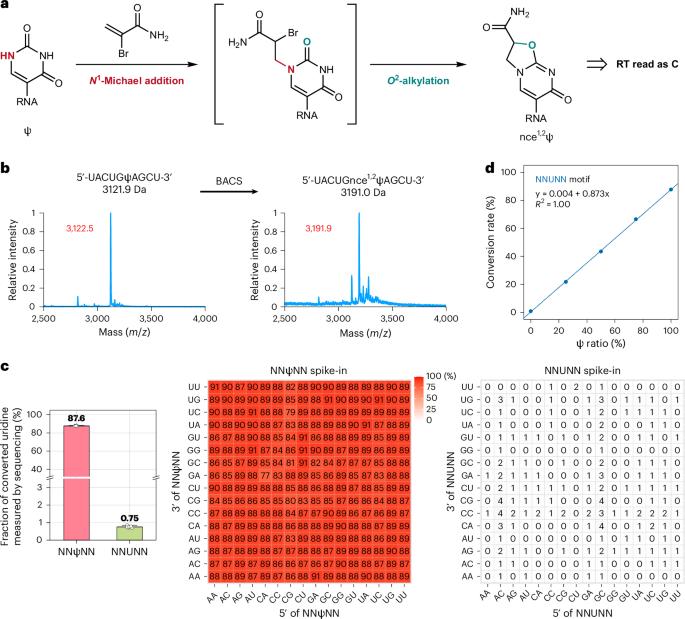
伪尿嘧啶(Ψ)是细胞 RNA 中最丰富的修饰之一。然而,主要由于缺乏高灵敏度和精确的检测方法,其功能仍然难以捉摸。在这里,我们引入了 2-溴丙烯酰胺辅助环化测序(BACS),它能实现Ψ到C的转变,以单碱基分辨率定量分析Ψ。BACS 能够精确鉴定Ψ位置,尤其是在密集修饰的Ψ区域和连续尿苷序列中。BACS 检测了人类 rRNA 和剪接体小核 RNA 中所有已知的 Ψ 位点,并生成了人类小核 RNA 和 tRNA 的定量 Ψ 图谱。此外,BACS 还能同时检测腺苷-肌苷编辑位点和 N1-甲基腺苷。假尿嘧啶合成酶 TRUB1、PUS7 和 PUS1 的消耗阐明了它们的靶标和序列基序。我们还在 Epstein-Barr 病毒编码的小 RNA EBER2 中发现了一个高含量的 Ψ114 位点。令人惊讶的是,将 BACS 应用于一组 RNA 病毒时,发现它们的病毒转录本或基因组中没有Ψ,从而揭示了不同病毒家族中假酰化的差异。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Absolute quantitative and base-resolution sequencing reveals comprehensive landscape of pseudouridine across the human transcriptome
Pseudouridine (Ψ) is one of the most abundant modifications in cellular RNA. However, its function remains elusive, mainly due to the lack of highly sensitive and accurate detection methods. Here, we introduced 2-bromoacrylamide-assisted cyclization sequencing (BACS), which enables Ψ-to-C transitions, for quantitative profiling of Ψ at single-base resolution. BACS allowed the precise identification of Ψ positions, especially in densely modified Ψ regions and consecutive uridine sequences. BACS detected all known Ψ sites in human rRNA and spliceosomal small nuclear RNAs and generated the quantitative Ψ map of human small nucleolar RNA and tRNA. Furthermore, BACS simultaneously detected adenosine-to-inosine editing sites and N1-methyladenosine. Depletion of pseudouridine synthases TRUB1, PUS7 and PUS1 elucidated their targets and sequence motifs. We further identified a highly abundant Ψ114 site in Epstein–Barr virus-encoded small RNA EBER2. Surprisingly, applying BACS to a panel of RNA viruses demonstrated the absence of Ψ in their viral transcripts or genomes, shedding light on differences in pseudouridylation across virus families. This study introduces a chemical method, BACS, that generates Ψ-to-C mutation signatures, allowing for sequencing and quantification of Ψ at single-base resolution.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


