微生物组-代谢物联系推动永冻土融化梯度上的温室气体动态变化
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
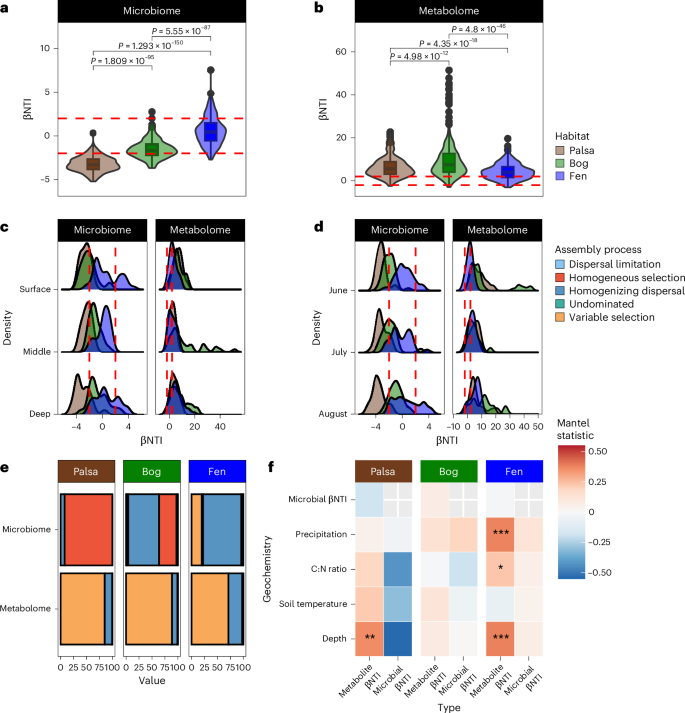
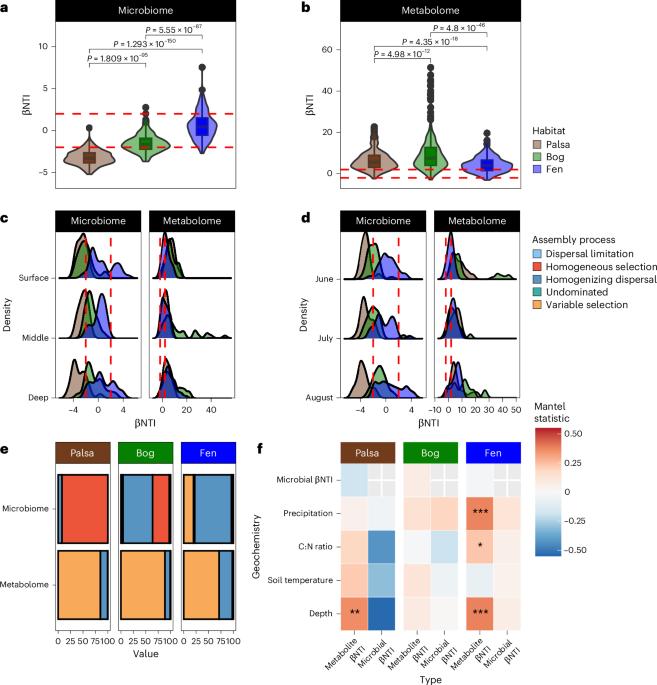
微生物组和代谢物之间的相互作用在环境中起着至关重要的作用,但这些相互作用如何在生态系统变化过程中推动温室气体排放仍不清楚。在这里,我们以群落组装理论为指导,利用成对的基因组分辨元基因组学和高分辨率傅立叶变换离子回旋共振质谱分析了瑞典 Stordalen Mire 永久冻土融化梯度上的微生物和代谢物组成,以检验微生物和代谢物是否对不断变化的驱动因素表现出一致的反应。我们的分析表明,推断出的微生物与代谢物组装过程之间存在差异,这表明它们对相同的选择性压力做出了不同的反应。这与基于性状的微生物模型中的常见假设相矛盾,并凸显了仅测量微生物群落级数据的局限性。此外,特征尺度分析揭示了微生物类群、代谢物与观测到的二氧化碳和甲烷孔隙水变化之间的联系。我们的研究展示了利用特征级数据和微生物与代谢物之间的相互作用来更好地理解生态系统变化过程中驱动温室气体排放的代谢过程所获得的洞察力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Microbiome–metabolite linkages drive greenhouse gas dynamics over a permafrost thaw gradient
Interactions between microbiomes and metabolites play crucial roles in the environment, yet how these interactions drive greenhouse gas emissions during ecosystem changes remains unclear. Here we analysed microbial and metabolite composition across a permafrost thaw gradient in Stordalen Mire, Sweden, using paired genome-resolved metagenomics and high-resolution Fourier transform ion cyclotron resonance mass spectrometry guided by principles from community assembly theory to test whether microorganisms and metabolites show concordant responses to changing drivers. Our analysis revealed divergence between the inferred microbial versus metabolite assembly processes, suggesting distinct responses to the same selective pressures. This contradicts common assumptions in trait-based microbial models and highlights the limitations of measuring microbial community-level data alone. Furthermore, feature-scale analysis revealed connections between microbial taxa, metabolites and observed CO2 and CH4 porewater variations. Our study showcases insights gained by using feature-level data and microorganism–metabolite interactions to better understand metabolic processes that drive greenhouse gas emissions during ecosystem changes. A multi-omics investigation at Sweden’s Stordalen Mire shows that microbial dynamics and metabolites must be taken into account to predict ecosystem responses to environmental change.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


