与 DNA 结合的 Spo11 核心复合物的冷冻电镜结构
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
启动减数分裂重组的 DNA 双链断裂是由拓扑异构酶相关酶 Spo11 在保守辅助因子的支持下形成的。由于尚未获得高分辨率的结构数据,有关 Spo11 及其伙伴的结构以及它们如何与 DNA 结合的许多问题依然存在。我们报告了酿酒酵母 Spo11 与 Rec102、Rec104 和 Ski8 的 DNA 结合核心复合物的高达 3.3 Å 分辨率的低温电子显微镜结构。在这些结构中,单体核心复合物与 DNA 主干以及凹陷的 3′-OH和第一个 5′悬垂核苷酸广泛接触,从而确定了 DNA 末端结合特异性的分子决定因素,并深入了解了体内 DNA 的裂解偏好。单个亚基及其界面的结构得到了酵母中功能数据的支持,使人们深入了解了金属离子在 DNA 结合中的作用,并发现了核心复合物 Top6BL 组成部分同源物中意想不到的结构变异。本文章由计算机程序翻译,如有差异,请以英文原文为准。
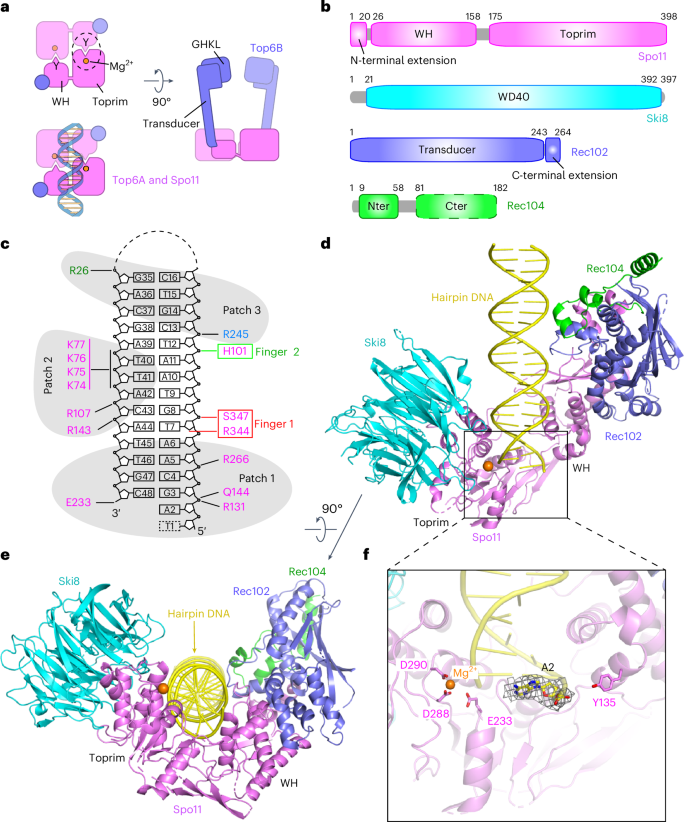

Cryo-EM structures of the Spo11 core complex bound to DNA
DNA double-strand breaks that initiate meiotic recombination are formed by the topoisomerase-relative enzyme Spo11, supported by conserved auxiliary factors. Because high-resolution structural data have not been available, many questions remain about the architecture of Spo11 and its partners and how they engage with DNA. We report cryo-electron microscopy structures at up to 3.3-Å resolution of DNA-bound core complexes of Saccharomyces cerevisiae Spo11 with Rec102, Rec104 and Ski8. In these structures, monomeric core complexes make extensive contacts with the DNA backbone and with the recessed 3′-OH and first 5′ overhanging nucleotide, establishing the molecular determinants of DNA end-binding specificity and providing insight into DNA cleavage preferences in vivo. The structures of individual subunits and their interfaces, supported by functional data in yeast, provide insight into the role of metal ions in DNA binding and uncover unexpected structural variation in homologs of the Top6BL component of the core complex. High-resolution structures of DNA-bound multiprotein Spo11 complexes, the DNA-cleaving ensembles that initiate meiotic recombination, elucidate the structural basis of its DNA-binding specificity and explain recombination initiation patterns in vivo.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


