通过单细胞转录组学系统分析细胞间通讯的 CellChat
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
单细胞测序技术的最新进展提供了一个机会,可以在减少偏差的情况下系统地探索组织中的细胞-细胞通讯。一个关键的挑战是将已知的分子相互作用和测量结果整合到一个框架中,以识别和分析复杂的细胞-细胞通讯网络。此前,我们开发了一种名为 CellChat 的计算工具,它能在一个易于解释的框架内从单细胞转录组数据中推断和分析细胞通讯网络。CellChat 采用基于质量作用的简化模型,将配体与多亚基结构受体之间的核心相互作用以及辅助因子的调制结合在一起,量化了两个细胞群之间的信号交流概率。重要的是,CellChat 利用各种定量指标和机器学习方法对细胞间通信进行了系统的比较分析。CellChat v2 是一个更新版本,包括更多的比较功能、配体-受体配对的扩展数据库以及丰富的功能注释和交互式 CellChat 浏览器。在这里,我们提供了在单细胞转录组数据上使用 CellChat v2 的分步方案,包括推断和分析来自一个数据集的细胞-细胞通讯,以及识别来自不同数据集、不同生物条件下改变的细胞间通讯、信号和细胞群。CellChat v2 工具包的 R 实现及其教程和图形输出可在 https://github.com/jinworks/CellChat 上获取。根据数据集的大小,该程序通常需要约 5 分钟,需要对 R 和单细胞数据分析有基本的了解,但不需要专门的生物信息学培训即可实现。本文章由计算机程序翻译,如有差异,请以英文原文为准。
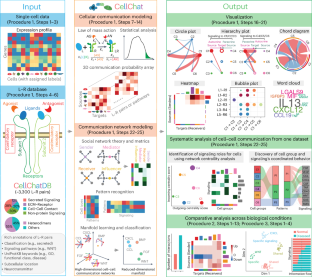

CellChat for systematic analysis of cell–cell communication from single-cell transcriptomics
Recent advances in single-cell sequencing technologies offer an opportunity to explore cell–cell communication in tissues systematically and with reduced bias. A key challenge is integrating known molecular interactions and measurements into a framework to identify and analyze complex cell–cell communication networks. Previously, we developed a computational tool, named CellChat, that infers and analyzes cell–cell communication networks from single-cell transcriptomic data within an easily interpretable framework. CellChat quantifies the signaling communication probability between two cell groups using a simplified mass-action-based model, which incorporates the core interaction between ligands and receptors with multisubunit structure along with modulation by cofactors. Importantly, CellChat performs a systematic and comparative analysis of cell–cell communication using a variety of quantitative metrics and machine-learning approaches. CellChat v2 is an updated version that includes additional comparison functionalities, an expanded database of ligand–receptor pairs along with rich functional annotations, and an Interactive CellChat Explorer. Here we provide a step-by-step protocol for using CellChat v2 on single-cell transcriptomic data, including inference and analysis of cell–cell communication from one dataset and identification of altered intercellular communication, signals and cell populations from different datasets across biological conditions. The R implementation of CellChat v2 toolkit and its tutorials together with the graphic outputs are available at https://github.com/jinworks/CellChat . This protocol typically takes ~5 min depending on dataset size and requires a basic understanding of R and single-cell data analysis but no specialized bioinformatics training for its implementation. CellChat enables systematic inference, quantitative analysis and intuitive visualization of cell–cell communication from single-cell transcriptomic data, as well as comparative analysis of intercellular communication across biological conditions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


