原生质谱法揭示了 SARS-CoV-2 PLpro 与抑制剂和细胞靶标的结合相互作用
IF 8.3
2区 材料科学
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
摘要
该蛋白酶通过影响病毒蛋白质的产生和拮抗宿主的抗病毒反应而在冠状病毒疾病中发挥关键作用。我们利用紫外光解离(UVPD)和变温电喷雾离子化(vT ESI)来定位 PLpro 抑制剂的结合位点,并揭示了抑制剂对蛋白质三级结构的稳定作用。我们比较了 SARS-CoV-1 和 SARS-CoV-2 的 PLpro 与抑制剂和 ISG15 的相互作用,以发现蛋白酶功能的可能差异。我们利用缺乏一个半胱氨酸的 PLpro 突变体来定位抑制剂的结合,热力学测量显示抑制剂 PR-619 稳定了折叠的 PLpro 结构。这些结果将为进一步开发PLpro作为抗击SARS-CoV-2和其他新出现的冠状病毒的治疗靶点提供信息。本文章由计算机程序翻译,如有差异,请以英文原文为准。
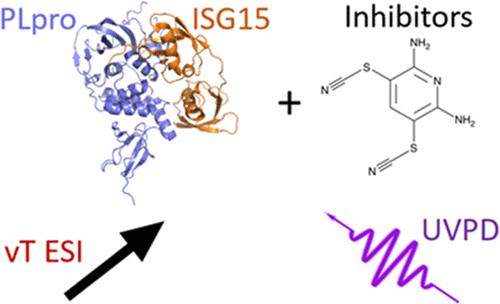
Native Mass Spectrometry Reveals Binding Interactions of SARS-CoV-2 PLpro with Inhibitors and Cellular Targets
Here we used native mass spectrometry (native MS) to probe a SARS-CoV protease, PLpro, which plays critical roles in coronavirus disease by affecting viral protein production and antagonizing host antiviral responses. Ultraviolet photodissociation (UVPD) and variable temperature electrospray ionization (vT ESI) were used to localize binding sites of PLpro inhibitors and revealed the stabilizing effects of inhibitors on protein tertiary structure. We compared PLpro from SARS-CoV-1 and SARS-CoV-2 in terms of inhibitor and ISG15 interactions to discern possible differences in protease function. A PLpro mutant lacking a single cysteine was used to localize inhibitor binding, and thermodynamic measurements revealed that inhibitor PR-619 stabilized the folded PLpro structure. These results will inform further development of PLpro as a therapeutic target against SARS-CoV-2 and other emerging coronaviruses.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
ACS Applied Materials & Interfaces
工程技术-材料科学:综合
CiteScore
16.00
自引率
6.30%
发文量
4978
审稿时长
1.8 months
期刊介绍:
ACS Applied Materials & Interfaces is a leading interdisciplinary journal that brings together chemists, engineers, physicists, and biologists to explore the development and utilization of newly-discovered materials and interfacial processes for specific applications. Our journal has experienced remarkable growth since its establishment in 2009, both in terms of the number of articles published and the impact of the research showcased. We are proud to foster a truly global community, with the majority of published articles originating from outside the United States, reflecting the rapid growth of applied research worldwide.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


