通过规范和非规范组蛋白解体实现核糖体 DNA 解旋的途径
IF 5.1
1区 生物学
Q1 BIOLOGY
引用次数: 0
摘要
包括哺乳动物特异性 H2A 变体 H2A.B 在内的核小体在精子发生、胚胎发育和肿瘤发生中发挥着关键作用,表明它在转录调控中的独特参与有别于典型的 H2A 核小体。尽管其意义重大,但确切的调控机制仍然难以捉摸。本研究利用固态纳米孔研究 DNA 的解旋动力学,在 DNA 和组蛋白之间施加局部作用力。对典型 H2A 和 H2A.B 核糖体的比较分析表明,H2A.B 变体需要较低的电压才能完全解开 DNA。此外,同步分析和分子动力学模拟表明,H2A.B 变体能迅速解开 DNA,使 H2A-H2B 二聚体在组蛋白八聚体解体后立即与 DNA 分离。与此相反,典型的 H2A 核小体解开 DNA 的速度较慢,这表明 H2A-H2B 二聚体在孔中处于堆积状态。这些研究结果表明,H2A.B核小体中的核糖体DNA经历了与典型H2A核小体不同的DNA解旋过程,其中涉及组蛋白八聚体的解体,从而使解旋过程更加顺畅。分子动力学模拟和纳米孔测量的综合方法有望发展成为研究分子相互作用的多功能工具,不仅可以研究核小体内部的相互作用,还可以通过强制解离 DNA 蛋白复合物进行研究。将分子动力学模拟和固态纳米孔测量相结合的研究揭示了不同的解体途径,这些途径可以解释 H2A.B 核小体在精子发生、胚胎发育和肿瘤发生过程中的不同特性和作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
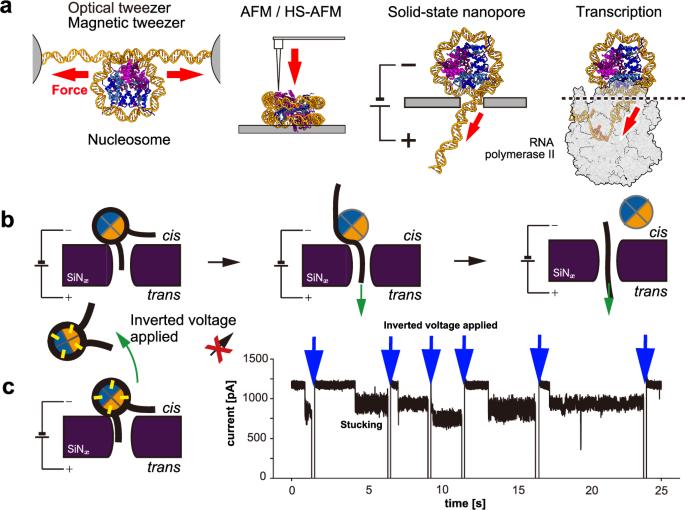
Nucleosomal DNA unwinding pathway through canonical and non-canonical histone disassembly
The nucleosome including H2A.B, a mammalian-specific H2A variant, plays pivotal roles in spermatogenesis, embryogenesis, and oncogenesis, indicating unique involvement in transcriptional regulation distinct from canonical H2A nucleosomes. Despite its significance, the exact regulatory mechanism remains elusive. This study utilized solid-state nanopores to investigate DNA unwinding dynamics, applying local force between DNA and histones. Comparative analysis of canonical H2A and H2A.B nucleosomes demonstrated that the H2A.B variant required a lower voltage for complete DNA unwinding. Furthermore, synchronization analysis and molecular dynamics simulations indicate that the H2A.B variant rapidly unwinds DNA, causing the H2A-H2B dimer to dissociate from DNA immediately upon disassembly of the histone octamer. In contrast, canonical H2A nucleosomes unwind DNA at a slower rate, suggesting that the H2A-H2B dimer undergoes a state of stacking at the pore. These findings suggest that nucleosomal DNA in the H2A.B nucleosomes undergoes a DNA unwinding process involving histone octamer disassembly distinct from that of canonical H2A nucleosomes, enabling smoother unwinding. The integrated approach of MD simulations and nanopore measurements is expected to evolve into a versatile tool for studying molecular interactions, not only within nucleosomes but also through the forced dissociation of DNA-protein complexes. Research combining molecular dynamics simulations and solid-state nanopore measurements reveals distinct disassembly pathways that may explain the distinct properties and roles of H2A.B nucleosomes in spermatogenesis, embryogenesis, and oncogenesis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


