细胞系特异性基因簇被 ZNF274 固定在抑制性核仁周围区室中
IF 11.7
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
核仁长期以来被称为核糖体生物发生的场所,其在塑造三维(3D)基因组组织方面的作用也日益得到认可。然而,基因组选定区域靶向核仁相关结构域(NADs)的机制仍然是个谜。在这里,我们揭示了ZNF274在NADs内隔离谱系特异性基因簇的重要作用,ZNF274是含锌指蛋白(KZFP)家族中的一个SCAN成员。消减 ZNF274 会引发整个基因组邻域的转录激活--其中包括原粘连蛋白和 KZFP 编码基因--抑制性染色质标记的缺失、三维基因组结构的改变以及新的 CTCF 结合。从机理上讲,ZNF274 将目标 DNA 序列锚定在核仁上,并通过 SCAN 结构域以前未知的功能促进其分隔。我们的研究结果阐明了 NAD 组织的基本机制,并表明核仁周围的抑制性中枢限制了串联排列基因的激活,从而在发育过程中实现选择性表达并调节细胞分化程序。本文章由计算机程序翻译,如有差异,请以英文原文为准。
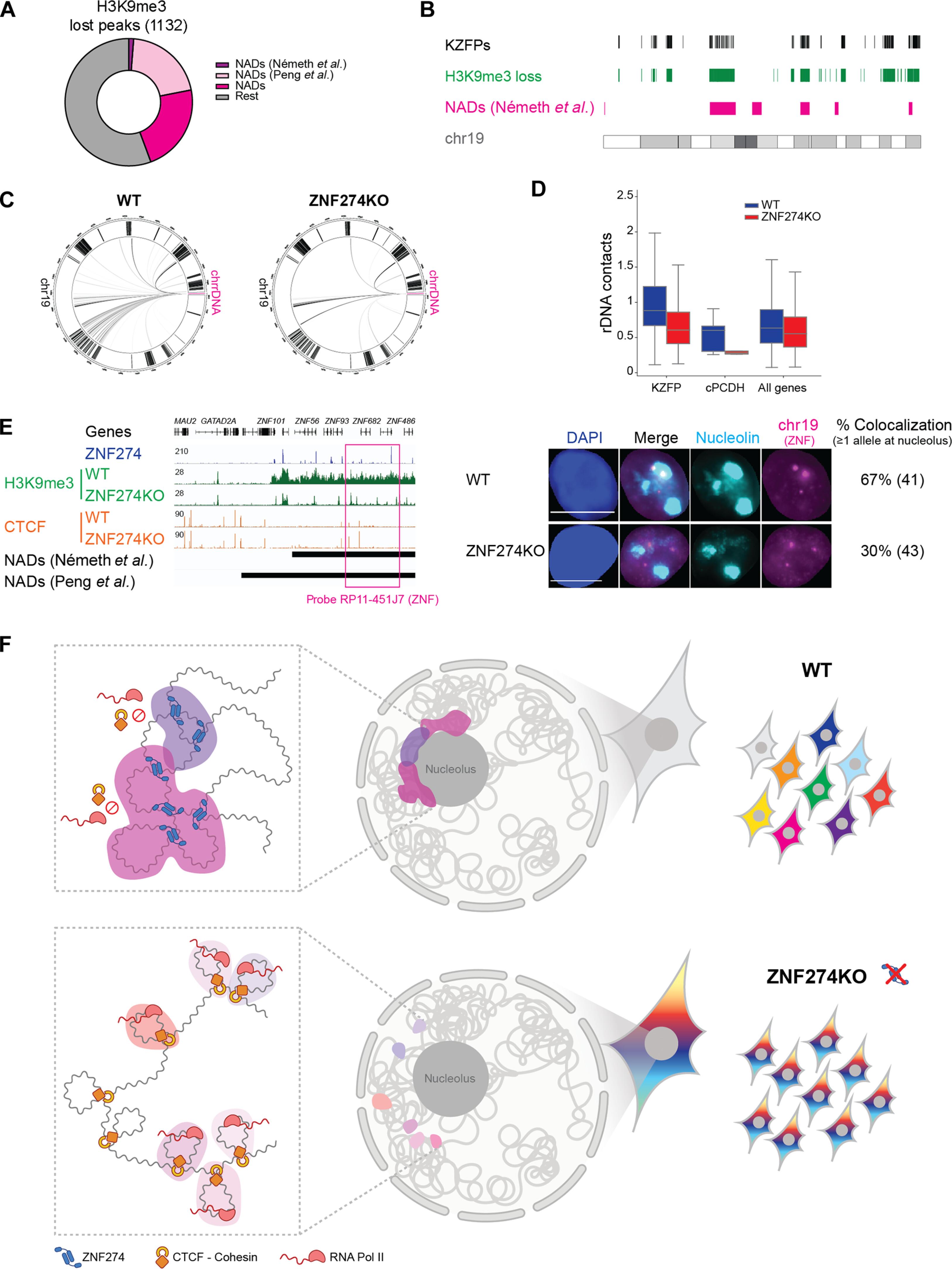
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Long known as the site of ribosome biogenesis, the nucleolus is increasingly recognized for its role in shaping three-dimensional (3D) genome organization. Still, the mechanisms governing the targeting of selected regions of the genome to nucleolus-associated domains (NADs) remain enigmatic. Here, we reveal the essential role of ZNF274, a SCAN-bearing member of the Krüppel-associated box (KRAB)–containing zinc finger protein (KZFP) family, in sequestering lineage-specific gene clusters within NADs. Ablation of ZNF274 triggers transcriptional activation across entire genomic neighborhoods—encompassing, among others, protocadherin and KZFP-encoding genes—with loss of repressive chromatin marks, altered the 3D genome architecture and de novo CTCF binding. Mechanistically, ZNF274 anchors target DNA sequences at the nucleolus and facilitates their compartmentalization via a previously uncharted function of the SCAN domain. Our findings illuminate the mechanisms underlying NAD organization and suggest that perinucleolar entrapment into repressive hubs constrains the activation of tandemly arrayed genes to enable selective expression and modulate cell differentiation programs during development.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


