IgE 浆细胞在转录和功能上有别于其他同型细胞。
IF 17.6
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
摘要
了解免疫球蛋白 E(IgE)产生细胞的表型和转录特征对于浆细胞(PC)生物学和过敏症治疗干预措施的开发至关重要。在这里,我们利用小鼠鼻内接触屋尘螨(HDM)的模型研究发现,在小鼠接触屋尘螨的早期(7 周),肺引流淋巴结(dLNs)中出现了短寿命的 IgE PCs。与其他 Ig 异型相比,IgE PCs 在这些不同组织中具有独特的表面和基因表达谱。IgE BMPCs上调了与前生存和BM归巢相关的基因,而IgE dLN PCs则表达了与最近的类别转换和分化相关的基因。与 IgG1 相比,IgE PCs 还表现出更高的内质网(ER)应激和蛋白编码基因表达,以及更高的抗体分泌率。总之,本研究强调了短寿命和长寿命 IgE PCs 独特的发育路径和转录特征。本文章由计算机程序翻译,如有差异,请以英文原文为准。
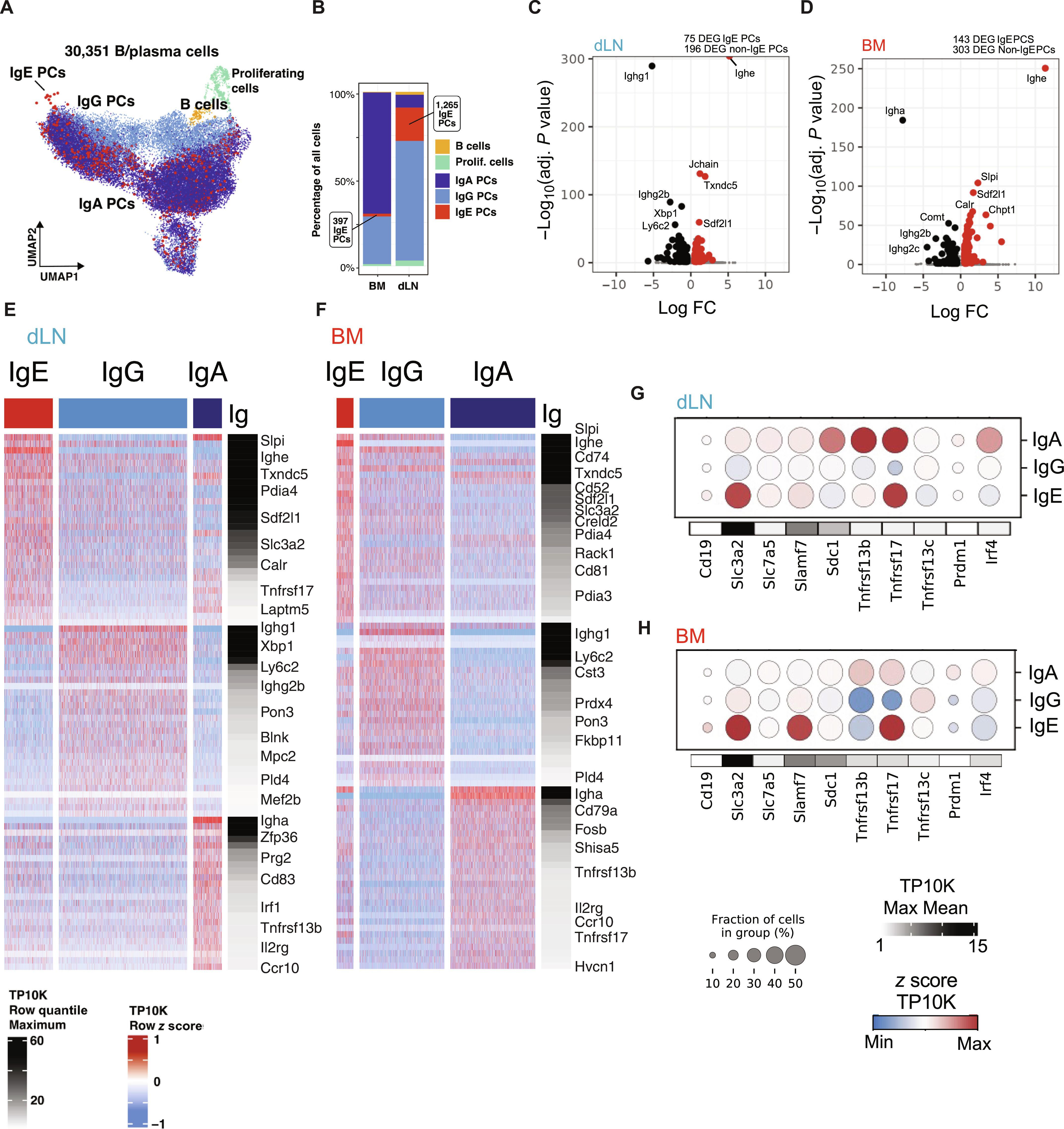
IgE plasma cells are transcriptionally and functionally distinct from other isotypes
Understanding the phenotypic and transcriptional signature of immunoglobulin E (IgE)–producing cells is fundamental to plasma cell (PC) biology and development of therapeutic interventions for allergy. Here, using a mouse model of intranasal house dust mite (HDM) exposure, we showed that short-lived IgE PCs emerge in lung draining lymph nodes (dLNs) during early exposure (<3 weeks) and long-lived IgE PCs accumulate in the bone marrow (BM) with prolonged exposure (>7 weeks). IgE PCs had distinct surface and gene expression profiles in these different tissues compared with other Ig isotypes. IgE BMPCs up-regulated genes associated with prosurvival and BM homing, whereas IgE dLN PCs expressed genes associated with recent class switching and differentiation. IgE PCs also exhibited higher expression of endoplasmic reticulum (ER) stress and protein coding genes and higher antibody secretion rate when compared with IgG1. Overall, this study highlights the unique developmental path and transcriptional signature of short-lived and long-lived IgE PCs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Immunology
Immunology and Microbiology-Immunology
CiteScore
32.90
自引率
2.00%
发文量
183
期刊介绍:
Science Immunology is a peer-reviewed journal that publishes original research articles in the field of immunology. The journal encourages the submission of research findings from all areas of immunology, including studies on innate and adaptive immunity, immune cell development and differentiation, immunogenomics, systems immunology, structural immunology, antigen presentation, immunometabolism, and mucosal immunology. Additionally, the journal covers research on immune contributions to health and disease, such as host defense, inflammation, cancer immunology, autoimmunity, allergy, transplantation, and immunodeficiency. Science Immunology maintains the same high-quality standard as other journals in the Science family and aims to facilitate understanding of the immune system by showcasing innovative advances in immunology research from all organisms and model systems, including humans.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


