优化的双子叶植物基因编辑技术可在番茄和拟南芥中实现可遗传的所需编辑
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
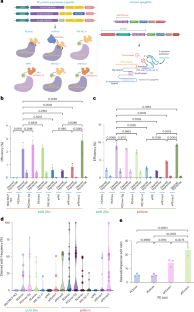
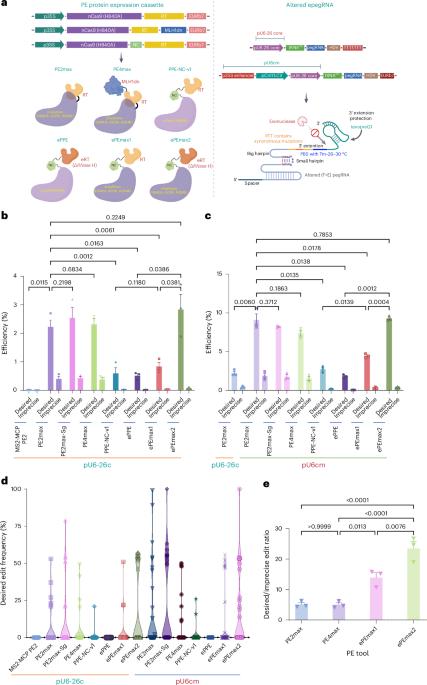
Prime editing(PE)可对动物和植物进行几乎所有类型的精确基因组编辑。该技术已成功应用于多种植物的基因组编辑,具有不同的效率和多功能性。然而,这种技术在双子叶植物中效率低下,原因不明。在这里,我们使用了新的PE元件组合,包括RNA伴侣和由PolII-PolIII复合启动子和病毒复制子系统驱动的经过改造的主编辑引导RNA,通过靶向深度测序评估,我们在胼胝体阶段获得了高达9.7%的预期PE效率。随后,我们发现在番茄和拟南芥中,高达 38.2% 的转化体含有所需的 PE 等位基因,这标志着成功的 PE 遗传传递。我们的 PE 工具还表现出了高准确性、特异性和复用能力,为双子叶植物中 PE 的实际应用挖掘了潜力,为植物科学的变革性进步铺平了道路。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Optimized dicot prime editing enables heritable desired edits in tomato and Arabidopsis
Prime editing (PE) enables almost all types of precise genome editing in animals and plants. It has been successfully adapted to edit several plants with variable efficiency and versatility. However, this technique is inefficient for dicots for unknown reasons. Here, using new combinations of PE components, including an RNA chaperone and altered engineered prime editing guide RNAs driven by a PolII–PolIII composite promoter and a viral replicon system, we obtained up to 9.7% of the desired PE efficiency at the callus stage as assessed by targeted deep sequencing. Subsequently, we identified that up to 38.2% of transformants contained desired PE alleles in tomatoes and Arabidopsis, marking successful heritable PE transmission. Our PE tools also showed high accuracy, specificity and multiplexing capability, which unlocked the potential for practical PE applications in dicots, paving the way for transformative advancements in plant sciences. This study enhanced prime editing (PE) for dicot plants using new combinations of PE components delivered by a geminiviral replicon. This achieved up to 38.2% PE efficiency in tomatoes and Arabidopsis, enabling precise breeding applications in dicots.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


