利用 DiMeLo-seq 测绘蛋白质与 DNA 的相互作用。
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
我们最近开发了定向甲基化长读程测序(DiMeLo-seq)技术,用于绘制全基因组蛋白质-DNA相互作用图谱。DiMeLo-seq 能够绘制单个 DNA 分子上多个相互作用位点的图谱,分析内源性 DNA 甲基化背景下的蛋白质结合情况,确定单倍型特异性蛋白质-DNA 相互作用,并绘制基因组重复区域中的蛋白质-DNA 相互作用图谱,而这些区域是短线程方法难以研究的。通过 DiMeLo-seq 技术,利用蛋白 A 将 Hia5 甲基转移酶系在抗体上,使感兴趣的蛋白质附近的腺嘌呤发生原位甲基化,然后利用长读程单分子 DNA 测序平台(如 Nanopore 测序平台)直接读出腺嘌呤甲基化,检测蛋白质与 DNA 的相互作用。在此,我们介绍了执行 DiMeLo-seq 的详细方案和实用指南。该方案可在新鲜、轻度固定或冷冻的细胞核上运行。原位靶向甲基化需要 1-2 天,文库制备需要 1-5 天,具体取决于所需的片段长度,Nanopore 测序需要 1-3 天,具体取决于所需的测序深度。该方案需要基本的分子生物学技能和设备,以及一台 Nanopore 测序仪。我们还提供了一个 Python 软件包 dimelo,用于分析 DiMeLo-seq 数据。本文章由计算机程序翻译,如有差异,请以英文原文为准。
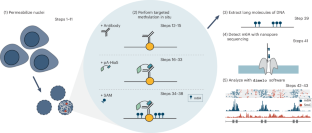
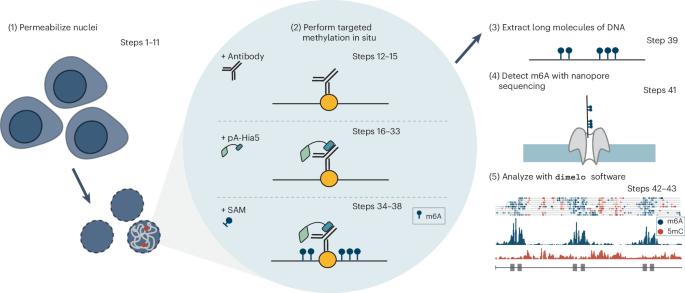
Mapping protein–DNA interactions with DiMeLo-seq
We recently developed directed methylation with long-read sequencing (DiMeLo-seq) to map protein–DNA interactions genome wide. DiMeLo-seq is capable of mapping multiple interaction sites on single DNA molecules, profiling protein binding in the context of endogenous DNA methylation, identifying haplotype-specific protein–DNA interactions and mapping protein–DNA interactions in repetitive regions of the genome that are difficult to study with short-read methods. With DiMeLo-seq, adenines in the vicinity of a protein of interest are methylated in situ by tethering the Hia5 methyltransferase to an antibody using protein A. Protein–DNA interactions are then detected by direct readout of adenine methylation with long-read, single-molecule DNA sequencing platforms such as Nanopore sequencing. Here we present a detailed protocol and practical guidance for performing DiMeLo-seq. This protocol can be run on nuclei from fresh, lightly fixed or frozen cells. The protocol requires 1–2 d for performing in situ targeted methylation, 1–5 d for library preparation depending on desired fragment length and 1–3 d for Nanopore sequencing depending on desired sequencing depth. The protocol requires basic molecular biology skills and equipment, as well as access to a Nanopore sequencer. We also provide a Python package, dimelo, for analysis of DiMeLo-seq data. DiMeLo-seq uses long-read, single-molecule sequencing to map protein–DNA interactions genome wide. This allows mapping of multiple interaction sites on single DNA molecules and profiling protein binding in the context of endogenous DNA methylation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


