利用三种 DNA 编码放大 FISH 成像策略观察单细胞中的表观遗传修饰及其空间邻近性:BEA-FISH、PPDA-FISH 和 Cell-TALKING。
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
核酸和蛋白质的表观遗传修饰及其空间邻近性在调节生理过程和疾病进展中发挥着重要作用。目前可用的细胞成像方法,如荧光原位杂交(FISH)和免疫荧光,很难检测到低丰度修饰及其空间邻近性。在此,我们将逐步介绍三种基于 DNA 编码放大 FISH 的成像策略,以克服这些挑战,满足不同的应用需求:碱基编码放大 FISH (BEA-FISH)、成对邻近分化放大 FISH (PPDA-FISH) 和细胞大分子系留 DNA 步行索引 (Cell-TALKING)。它们都使用类似的 DNA 编码扩增核心原理,将不同的非序列分子特征转化为独特的 DNA 条形码,用于原位滚圆扩增和 FISH 分析。这涉及固定细胞样本中的三个关键反应:靶标标记、DNA 编码和滚动圈扩增成像。利用该方案,这三种成像策略可分别实现低丰度修饰的原位计数、两种修饰的成对临近差异可视化以及一种蛋白质周围多种修饰的探索(一对多临近)。低丰度修饰,包括 5-羟甲基胞嘧啶、5-甲酰基胞嘧啶、5-羟甲基尿嘧啶和 5-甲酰基尿嘧啶,在单细胞中清晰可见。可发现核酸修饰和/或组蛋白修饰的各种组合模式。根据不同的成像应用,整个过程大约需要 2-4 天。本文章由计算机程序翻译,如有差异,请以英文原文为准。
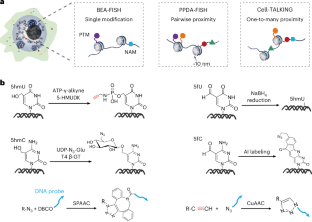

Visualizing epigenetic modifications and their spatial proximities in single cells using three DNA-encoded amplifying FISH imaging strategies: BEA-FISH, PPDA-FISH and Cell-TALKING
Epigenetic modifications and spatial proximities of nucleic acids and proteins play important roles in regulating physiological processes and disease progression. Currently available cell imaging methods, such as fluorescence in situ hybridization (FISH) and immunofluorescence, struggle to detect low-abundance modifications and their spatial proximities. Here we describe a step-by-step protocol for three DNA-encoded amplifying FISH-based imaging strategies to overcome these challenges for varying applications: base-encoded amplifying FISH (BEA-FISH), pairwise proximity-differentiated amplifying FISH (PPDA-FISH) and cellular macromolecules-tethered DNA walking indexing (Cell-TALKING). They all use the similar core principle of DNA-encoded amplification, which transforms different nonsequence molecular features into unique DNA barcodes for in situ rolling circle amplification and FISH analysis. This involves three key reactions in fixed cell samples: target labeling, DNA encoding and rolling circle amplification imaging. Using this protocol, these three imaging strategies achieve in situ counting of low-abundance modifications alone, the pairwise proximity-differentiated visualization of two modifications and the exploration of multiple modifications around one protein (one-to-many proximity), respectively. Low-abundance modifications, including 5-hydroxymethylcytosine, 5-formylcytosine, 5-hydroxymethyluracil and 5-formyluracil, are clearly visualized in single cells. Various combinatorial patterns of nucleic acid modifications and/or histone modifications are found. The whole protocol takes ~2–4 d to complete, depending on different imaging applications. The three methods present visualize epigenetic modifications and their spatial proximities in single cells; base-encoded amplifying FISH, pairwise proximity-differentiated amplifying FISH and cellular macromolecules-tethered DNA walking indexing.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


